Software: Difference between revisions
| Line 126: | Line 126: | ||
=====PhotoArtMaster===== | =====PhotoArtMaster===== | ||
Early versions of PhotoArtMaster are still '''available from Amazon''' at low prices (I'm not sure where they come from.) | Early versions of PhotoArtMaster are still '''available from Amazon''' at low prices (I'm not sure where they come from.) | ||
[http://www.amazon.co.uk/s/ref=nb_sb_noss?url=search-alias%3Daps&field-keywords=photoartmaster] . Some help for both the early versions and the latest version can be found in [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/SieveWebPages/a4a_2_screensize.pdf <span style="color: Chocolate">''''this | [http://www.amazon.co.uk/s/ref=nb_sb_noss?url=search-alias%3Daps&field-keywords=photoartmaster] . Some help for both the early versions and the latest version can be found in [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/SieveWebPages/a4a_2_screensize.pdf <span style="color: Chocolate">''''this document''''' </span>]). | ||
====The final version of the Windows ArtMaster2.0 [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/OpenSource_ArtMaster/ArtMaster2.0Release.zip <span style="color: #B31B1B">'''''is downloadable here'''''</span>] with no support.==== | ====The final version of the Windows ArtMaster2.0 [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/OpenSource_ArtMaster/ArtMaster2.0Release.zip <span style="color: #B31B1B">'''''is downloadable here'''''</span>] with no support.==== | ||
Unzip into (for example) the "Program Files" directory then set your system environment to include: ''C:\Program Files\Pam2.0 Release\jre\bin;C:\Program Files\Pam2.0 Release\bin;'' (You may need help for this. I right clicked 'computer' from the 'Start' menu, then selected 'Advanced system settings', then 'Environment Variables' and finally slid through the System variables until I found and selected 'Path'. This allowed me to edit the path by adding ';C:\Program Files\Pam2.0 Release\jre\bin;C:\Program Files\Pam2.0 Release\bin' to the end). Some sort of help using the software is available in [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/SieveWebPages/a4a_2_screensize.pdf <span style="color: Chocolate">''''this essay''''' </span>].<br><br> | Unzip into (for example) the "Program Files" directory then set your system environment to include: ''C:\Program Files\Pam2.0 Release\jre\bin;C:\Program Files\Pam2.0 Release\bin;'' (You may need help for this. I right clicked 'computer' from the 'Start' menu, then selected 'Advanced system settings', then 'Environment Variables' and finally slid through the System variables until I found and selected 'Path'. This allowed me to edit the path by adding ';C:\Program Files\Pam2.0 Release\jre\bin;C:\Program Files\Pam2.0 Release\bin' to the end). Some sort of help using the software is available in [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/SieveWebPages/a4a_2_screensize.pdf <span style="color: Chocolate">''''this essay''''' </span>].<br><br> | ||
Revision as of 10:36, 14 April 2014
Current activity: a collaboration with the CoenLab with the aim of understanding how patterns of gene activity in biological organs influence the developing shape. The BanghamLab is focussed on the conceptual underpinning: concepts captured in computational growth models, experimental data visualisation and analysis.
Notes on documenting our software
Notes for Lab members on how to contribute to this Wiki and where to put downloads.
Matlab tip: searching a large data structure for a particular field. Clear the command window. Evaluate the structure to list all the fields, then use the usual control-f search tool on the command window.
Computational biology
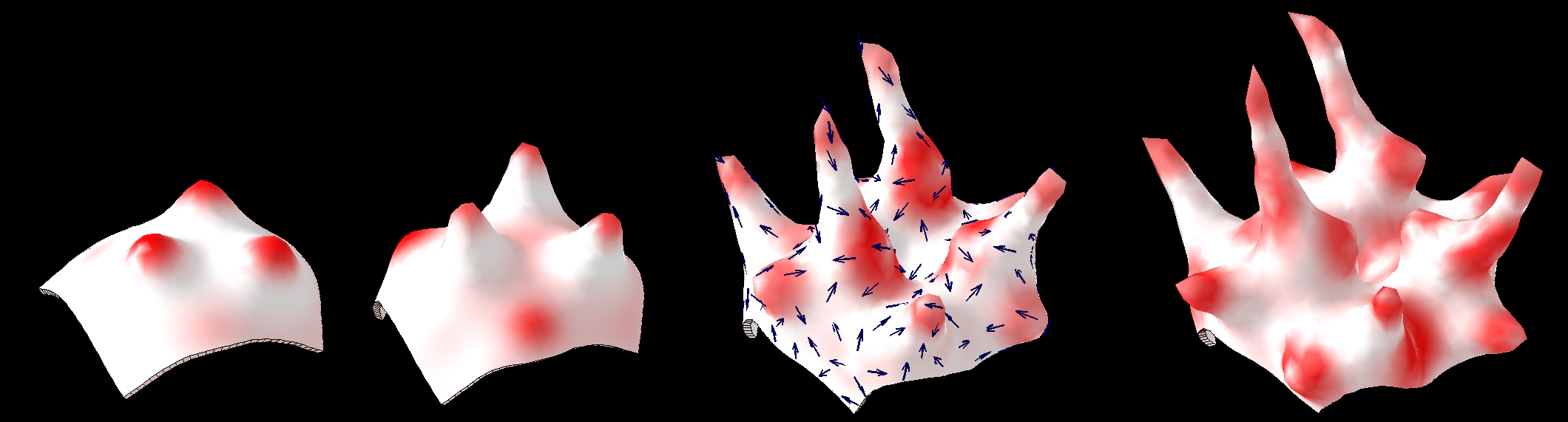
Quantitative understanding of growing shapes: GFtbox
We developed GFtbox to allow us to model the growth of complex shapes. The goal was and remains: to understand the relationships between genes, growth and form.
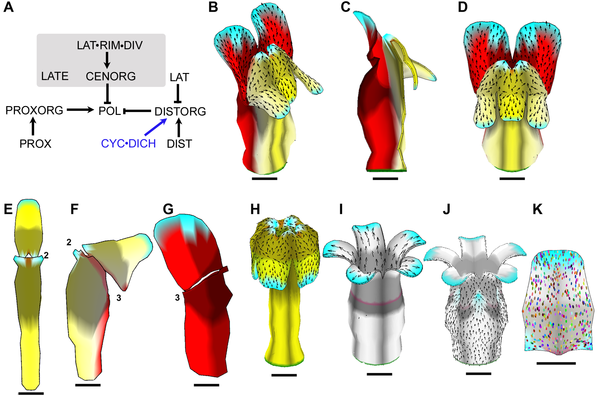 Example of a growing snapdragon flower and some mutants ( Green et al 2011). Growth is specified by factors (genes) according to the Growing Polarised Tissue Framework. Colours represent putative gene activity, arrows the polariser gradient and spots clones. Example of a growing snapdragon flower and some mutants ( Green et al 2011). Growth is specified by factors (genes) according to the Growing Polarised Tissue Framework. Colours represent putative gene activity, arrows the polariser gradient and spots clones.
|
GPT_Snapdragon_2010_Green_et_al-0002.png</wikiflv> |
| <imgicon>GPT_thumbnail2.png|120px|GFtbox</imgicon> |
For modelling the growth of shapes. Ready Reference Manual
What? How? Where? Background Download GFTbox from SourceForge Petals Sauret-Güeto et al 2013 |
GFtbox is an implementation of the Growing Polarised Tissue Framework (GPT-framework) for understanding and modelling the relationship between gene activity and the growth of shapes such leaves, flowers and animal embryos (Kennaway et al 2011). The GPT-framework was used to capture an understanding of (to model) the growing petal (Sauret-Güeto et al 2013), leaf (Kuchen et al 2012) and Snapdragon flower Green et al 2011. The Snapdragon model was validated by comparing the results with other mutant and transgenic flowers Cui et al 2010. The key point is how outgrowths can be specified by genes. The icon shows an asymmetrical outgrowth. Conceptually, it is specifed by two independent patterns under genetic control: a pattern of growth and a pattern of organisers. The outgrowth arises from a region of extra overall growth. Growth is aligned along axes set by two interacting systems. Organisers at the ends of the mesh create a lengthwise gradient. This gradient interacts with the second due to an organiser that generates polariser in a region that becomes the tip of the outgrowth. (Kennaway et al 2011) |
Viewing and measuring volume images: VolViewer
| <imgicon>VolViewer-logo.png|120px|VolViewer</imgicon> | For viewing and measuring volume images on both normal and stereo screens. Typical images from: confocal microscope and Optical Projection Tomography (OPT) images What? How? Where? Cell: Online Gallery | Front cover: Handbook of Plant Science | Front cover: The Plant Cell | American Scientist | Royal Microscopical Society: Infocus Magazine | Bundled with the Bioptonic 3001 scanner: Bioptonics Viewer | The Daily Mail | The Guardian newspaper: 3D Fruit fly | Qt Ambassador program | Triffid Nurseries website
|
VolViewer is used as a stand-alone app. or as a viewport for other systems, e.g. Matlab programs. VolViewer uses OpenGL and Qt to provide a user friendly application to interactively explore and quantify multi-dimensional biological images. It has been successfully used in our lab to explore and quantify confocal microscopy and optical projection tomography images. Written by Jerome Avondo it is open-source and is also compatible with the Open Microscopy Environment (OME) (Chris Allen and Avondo, et. al. OMERO: flexible, model-driven data management for experimental biology Nature Methods 9, 245–253 (2012)) ). ).
|
Analysing shapes in 2D and 3D: AAMToolbox
| <imgicon>AAMToolbox_logo.jpg|120px|AAMToolbox</imgicon> | For analysing populations of shapes and colours within the shapes using principal component analysis. What? How? Where?
|
The AAMToolbox enables the user analyse the shape and colour of collections of similar objects. Originally developed to analyse face shapes for lipreading (Matthews et al. 2002version of pdf), we have used it extensively for analysing the shapes of leaves (Langlade et al 2005.,Bensmihen et al. 2010) and petals (Whibley et al 2006,Feng et al. 2010). The analysis can be applied to art, for example, finding systematic differences between portraits by Rembrandt and Modigliani. |
Analysing the shapes of clones: SectorAnalysisToolbox
| <imgicon>Sector analysis icon.jpg|120px|SectorAnalysisToolbox</imgicon> | For analysing the shapes of marked cell clones. What? How? Where? |
The SectorAnalysisToolbox enables the user analyse the shapes of marked clones in a sheet of tissue. |
Algorithms
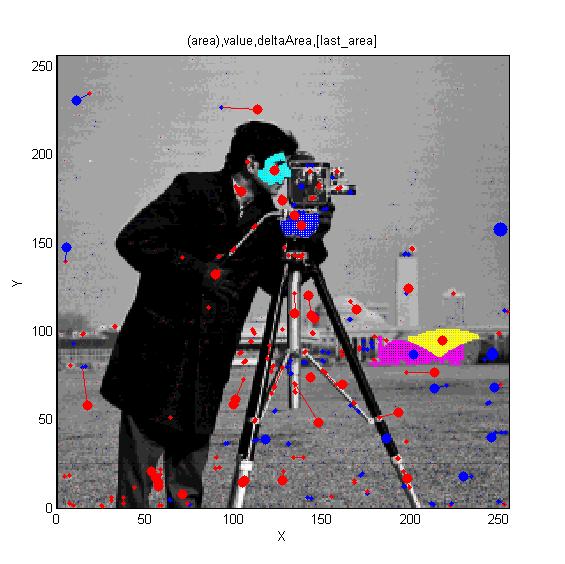
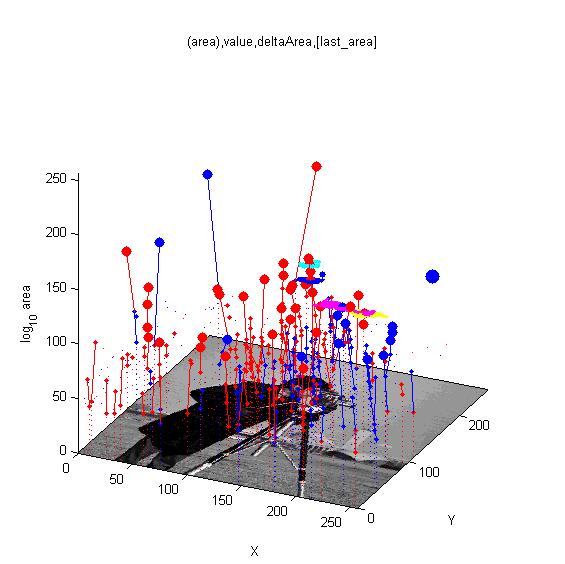
MSERs, extrema, connected-set filters and sieves
The algorithm finding MSER's starts with a connected-set opening or 'o' sieve
For finding features in and segmenting images.
What? How? Where?
Tutorials: from the beginning
Art, extrema of light and shade: PhotoArtMaster
Art created using ArtMaster was featured in an exhibit at the London Victoria and Albert (V&A) Museum exhibition 'Cheating? How to make Perfect Work of Art' (2003). The exhibition centered on the idea of Hockney's that advances in realism and accuracy in the history of Western art since the Renaissance were primarily the result of optical aids such as the camera obscura, camera lucida, and curved mirrors. My exhibit used a touch screen (rare in those days) and ArtMaster to help users create 'paintings'. finding its name.
 
|
PhotoArtMaster
Early versions of PhotoArtMaster are still available from Amazon at low prices (I'm not sure where they come from.) [1] . Some help for both the early versions and the latest version can be found in 'this document ).
The final version of the Windows ArtMaster2.0 is downloadable here with no support.
Unzip into (for example) the "Program Files" directory then set your system environment to include: C:\Program Files\Pam2.0 Release\jre\bin;C:\Program Files\Pam2.0 Release\bin; (You may need help for this. I right clicked 'computer' from the 'Start' menu, then selected 'Advanced system settings', then 'Environment Variables' and finally slid through the System variables until I found and selected 'Path'. This allowed me to edit the path by adding ';C:\Program Files\Pam2.0 Release\jre\bin;C:\Program Files\Pam2.0 Release\bin' to the end). Some sort of help using the software is available in 'this essay .
The sieve algorithms underpinning PhotoArtMaster software are described in an extract of the
'essay . These documents were written to support our Fo2Pix company. PhotoArtMaster originally sold >65,000 licences but ill health forced the closure of Fo2Pix.
|
Reaction-diffusion and morphogenesis
Illustration of morphogenesis inspired by Turing's paper.
Example using growth toolbox GPT_ReactionDiffusionTentacles_20121211.zip
1 A
| <wikiflv width="300" height="300" logo="false" loop="true" background="white">GPT_rd_rk_tentacles_20120417-0004.flv|GPT_rd_rk_tentacles_20120417-0004_First.png</wikiflv> A simple reaction-diffusion system develops a pattern of spots. | GPT_rd_rk_tentacles_20120417-0004.png</wikiflv>
|
Open source systems to which we contribute
OMERO
| <imgicon>OMERO_DIAGRAM.jpg|100px|OMERO</imgicon> | For working with the OME image database. See Details, Download OMERO Workshop (Windows, Mac, Linux) |
Open Microscopy Environment Remote Objects (OMERO). for visualising, managing, and annotating scientific image data. See also our OMERO Workshop training course we ran in April 2011. |
Tools and Utilities
BioformatsConverter
| <imgicon>BioformatsConverterZip.png|100px|BioformatsConverter</imgicon> | For converting microscope manufacturer proprietary file formats. See Details (Windows, Mac, Linux) |
This tool allows for the batch conversion of microscope manufacturer proprietary file formats, to the open source OME-TIFF standard. Uses the Bioformats library. |
Dependency Checking Tool
Tool for recursively finding what further functions a function depends on. See Details
In development
MTtbox
| <imgicon>MTtboxA.jpg|100px|BioformatsConverter</imgicon> | For modelling the behaviour of microtubules within a cell. See Details |
In development. The idea is to be able to model the behaviour of growing microtubules and factors as they react chemically and diffuse within the different cell compartments. The icon shows a spherical cell sliced open to show concentric components: cell wall (magenta), plasma-membrane (yellow), cytoplasm (green) and vacuole (yellow). Microtubules (blue) grow in 3D within the cytoplasm. |