VolViewer: Difference between revisions
JeromeAvondo (talk | contribs) No edit summary |
JeromeAvondo (talk | contribs) No edit summary |
||
| Line 23: | Line 23: | ||
* [[VolViewerUserManual|Quick Guide with exercises.]] | * [[VolViewerUserManual|Quick Guide with exercises.]] | ||
* [[VolViewerTutorials|Video Tutorials.]] | |||
=Image Gallery / Sample Data= | =Image Gallery / Sample Data= | ||
| Line 60: | Line 61: | ||
Public SVN: [https://cmpdartsvr1.cmp.uea.ac.uk/banghamlabSVN/VolViewer/ https://cmpdartsvr1.cmp.uea.ac.uk/banghamlabSVN/VolViewer/] | Public SVN: [https://cmpdartsvr1.cmp.uea.ac.uk/banghamlabSVN/VolViewer/ https://cmpdartsvr1.cmp.uea.ac.uk/banghamlabSVN/VolViewer/] | ||
=Media/Press= | =Media/Press= | ||
Revision as of 14:01, 24 October 2011
Description
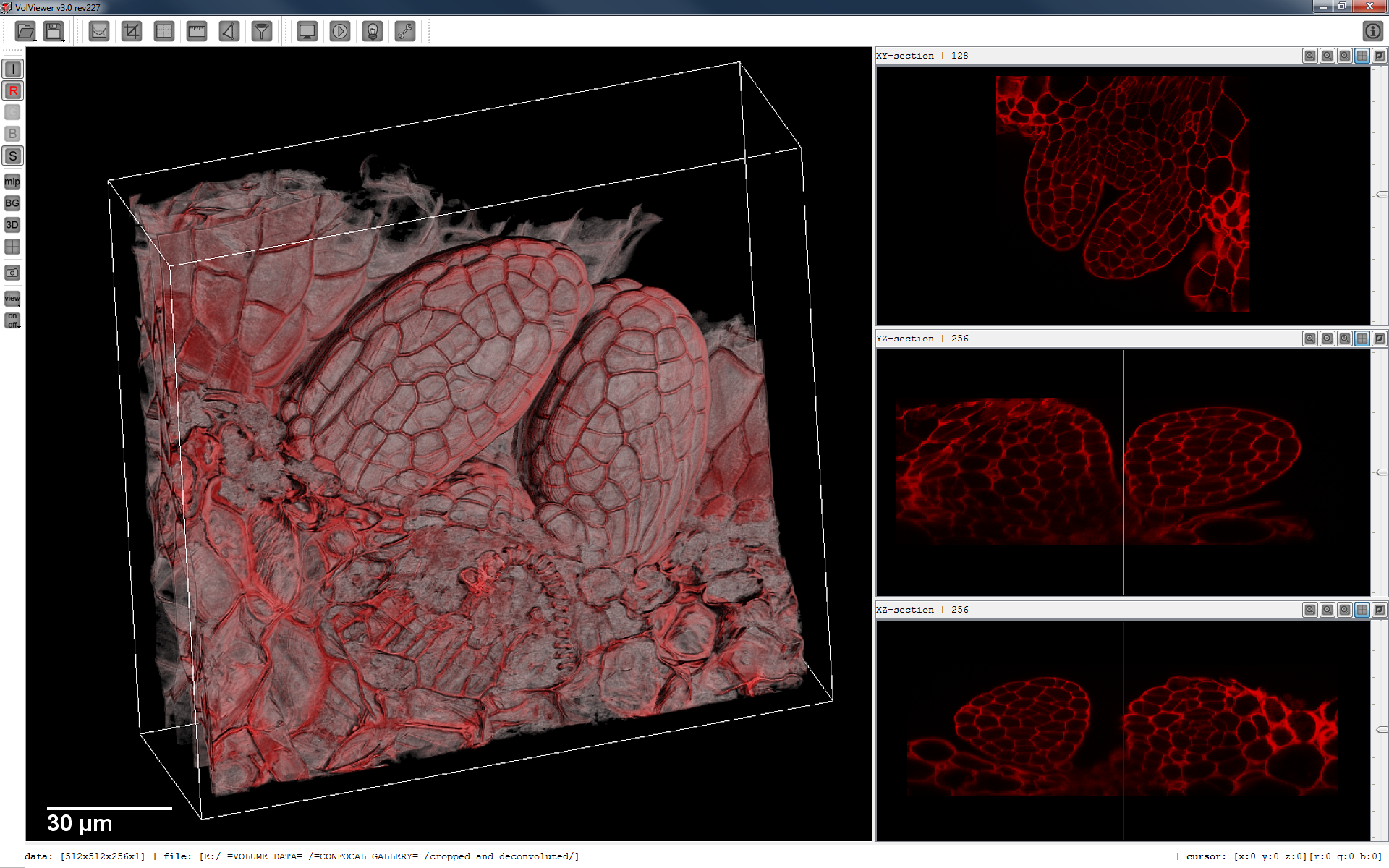
VolViewer is an open source application for the interactive visualisation and quantification of biological data. The application is written in C++ using OpenGL and Qt.
Features:
- Real-time volume rendering using an optimized 3D texture slicing algorithm.
- Interactive transfer functions to independently adjust opacity and luminance for up to three data channels.
- Real-time per channel thresholding, brightness and contrast operators.
- On-the-fly gradient computation for local illumination.
- Iso-surface computation with surface smoothing.
- Section viewing in any orientation / position.
- Real-time volume clipping.
- 3D measurements, filters & segmentation.
- Key frame interpolation for easy movie creation/export by interpolating transformations, clipping and transfer function states.
- Stereo rendering using either quad buffer or anaglyph mode.
Requirements: An OpenGL 2.1 / GLSL 1.20 compatible GPU with a recomended 512MB of memory.
User Manual
Image Gallery / Sample Data
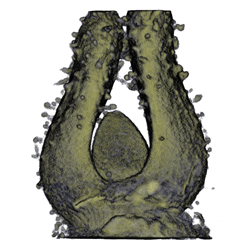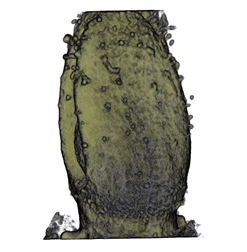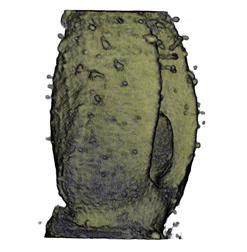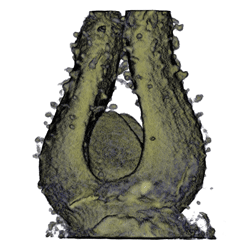 |
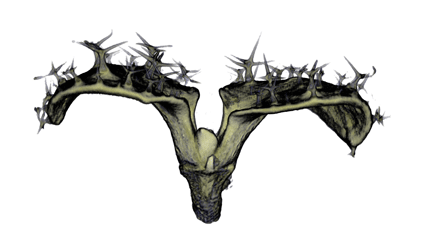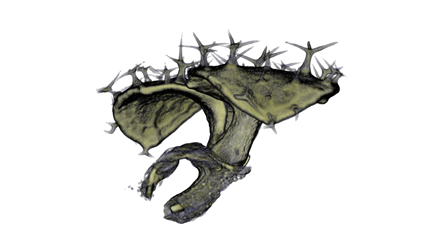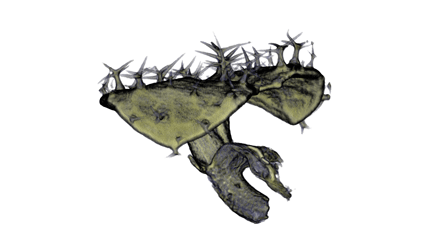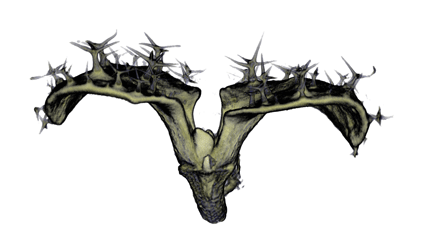 |
 |

|
| Antirinhium Meristem | Arabidopsis Seedling | Arabidopsis Leaf (GL2:GUS expression in red) | Arabidopsis Leaf (Ath8:::GUS expression in red) |
| Download | Download | Download | Download |
* all data courtesy of Karen Lee [1]
Download
Although we try to keep up to date builds these sometimes lag behind the SVN trunk. So if you want the latest version / features, it is best to build the application from the trunk of the SVN. The build system is based on qmake for easy cross platform compilation.
| Windows (32bit) | Windows (64bit) | Linux (64bit) | MacOS X (i386/x86_64/10.5+) |
OMERO Support
It is possible to build VolViewer with support for the Open Microscopy Environment, please see the guide below:
Source Code
Public SVN: https://cmpdartsvr1.cmp.uea.ac.uk/banghamlabSVN/VolViewer/
Media/Press
VolViewer has appeared in the following:
- Front cover: Handbook of Plant Science
- Front cover: Plant Cell September 2006
- Royal Microscopical Society: Infocus September 2009
- Bundled with the Bioptonic 3001 scanner: Bioptonics Viewer
- In the Guardian newspaper: 3D Fruit fly
- Showcased in the Qt Ambassador program.
- Showcased on the Triffid Nurseries website.