Back to BanghamLab software
What? How? Where?
What? We wish to understand how patterns of gene activity in biological organs influence the developing shape. The shapes arise because different regions of, for example a sheet of cells, grow at different rates. The problem is that a particular shape outline could arise through a number of different patterns of growth inside the outline. To understand growth it is, therefore, necessary to observe and measure growth rates throughout the growing organ. Small organs growth can be tracked at the cell scale using confocal microscopy. As the organ gets larger, this becomes impractical and we use an alternative: clonal analysis.
Clones of cells arise from divisions of a single cell. Clones become useful if the cell (and its daughters) are marked in some way. The mark could be the shape of hairs in a fly wing, or the colour of cells in a flower petal. As result of multiple cell divisions, growth, the shape of the clone (sometimes call the sector) can be followed by measuring the shape of the marked clone. The Sector Analysis Toolbox (SAT) is used to quantify patterns of growth by analysing the shapes of these marked sectors or clones.
|
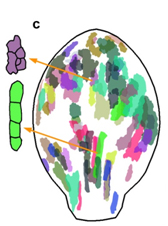
Individual clones observed using confocal microscopy in two regions of an Arabidopsis leaf together with clones (from a number of leaves) that have been identified, labelled and warped to the average leaf shape.
|
How? What does SectorAnalysisToolbox require?
Sector Analysis Toolbox is written in Matlab. It does not require any extra Mathworks toolboxes, nor any separately compiled modules. Matlab is available as a 30 day free trial and as a student edition. Sector Analysis Toolbox comprises around 20,000 lines of code.
Where?
Download