Research: Difference between revisions
(Replaced content with " Recent software<br><br> Publications<br><br> [http://www.uea.ac.uk/computing/People/Honorary/Andrew+Bangham Andrew Bangham at the Univers...") |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Software | | =[http://www.uea.ac.uk/computing/People/Honorary/Andrew+Bangham Bangham at UEA]= | ||
[[ | =<span style="color:Indigo;">Bangham Lab - Home= | ||
[http://www.uea.ac.uk/ | |||
<span style="color: DarkGreen">'''Current activity: a collaboration''' with the [http://rico-coen.jic.ac.uk/index.php/Main_Page CoenLab] with the aim of understanding how patterns of gene activity in biological organs influence the developing shape. The BanghamLab is focussed on the conceptual underpinning: concepts captured in computational growth models, experimental data visualisation and analysis.</span> | |||
=<span style="color:DarkGreen;">Computational biology toolboxes= | |||
---- | |||
==<span style="color:DarkGreen;">Growing complex biological shapes from patterns of gene expression</span>== | |||
{| border="0" width=100% style="background-color:#000000;" | |||
|- | |||
|align="center"| | |||
[[Image:LabelledCropped GPT Snapdragon 2010-000340-0001.png|120px]] | |||
[[Image:LabelledCropped GPT Snapdragon 2010-000490-0001.png|120px]] | |||
[[Image:LabelledCropped GPT Snapdragon 2010-000570-0002.png|120px]] | |||
[[Image:LabelledCropped GPT Snapdragon 2010-000570-0007.png|120px]] | |||
[[Image:LabelledCropped GPT Snapdragon 2010-000570-0003 double.png|100px]] | |||
[[Image:LabelledCropped GPT Snapdragon 2010-000570-0002 triple.png|120px]] | |||
|} | |||
<br> | |||
[[Software#Quantitative understanding of growing shapes: GFtbox|<span style="color:Green;">'''MORE'''</span>]]<br> | |||
Movies: [http://www.youtube.com/watch?v=7Uu5dHxyEwk Model Snapdragon flower movie], [http://www.youtube.com/watch?feature=endscreen&v=kZ45R1UlohM&NR=1 Why Snapdragon flower shape is so complex]<br><br> | |||
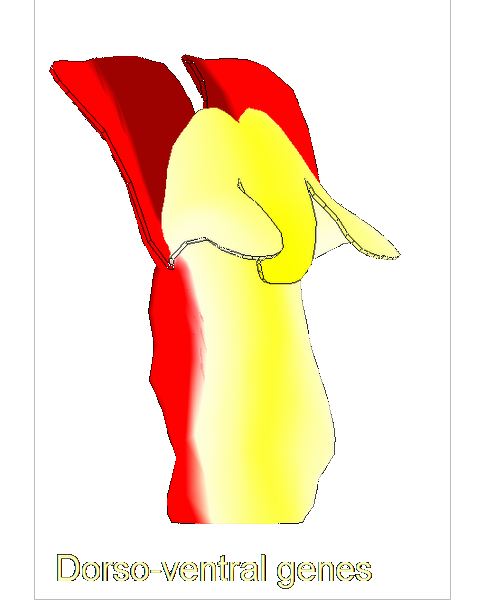
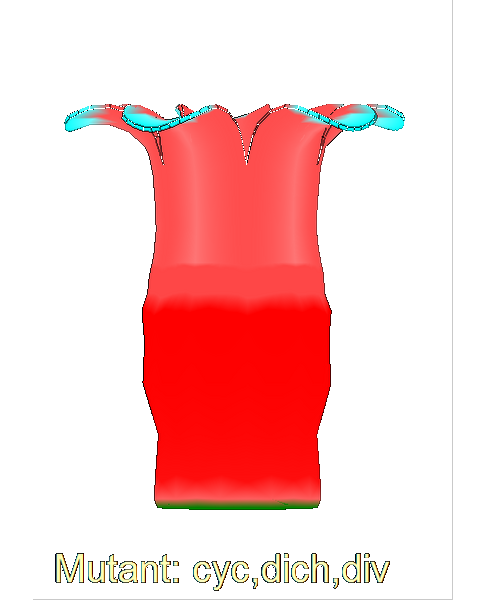
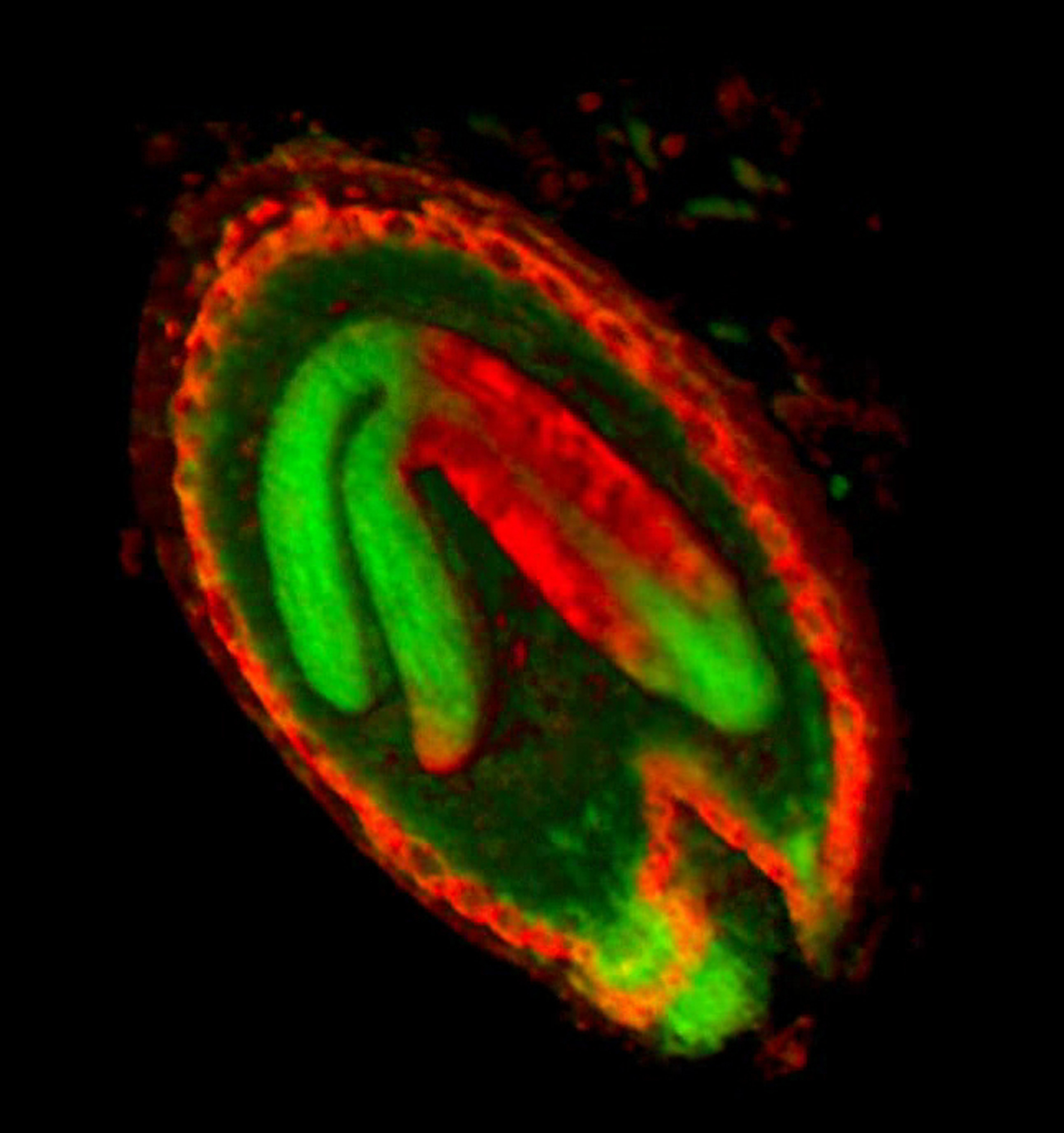
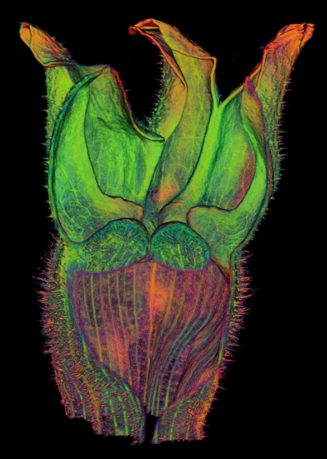
The growth of a complex ''snapdragon flower shape''. Key to the model, is an hypothesis on'' how organisers control the axes'' along which growth occurs. The organisers are shown in cyan and green. On the right are the shapes of two symmetrical mutants computed from the same model (hypotheses).<br><br> | |||
The Growing Polarised Tissue Framework for understanding and modelling the relationship between gene activity and the growth of shapes such leaves, flowers and animal embryos is introduced in ([http://www.ploscompbiol.org/article/info:doi/10.1371/journal.pcbi.1002071 Kennaway et al 2011]). The GPT-framework was used to capture an understanding of (to model) the Snapdragon flower [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1000537 Green et al 2011]. The Snapdragon model was validated by comparing the results with other mutant and transgenic flowers [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1000538 Cui et al 2010.]<br><br> | |||
The GPT-framework was also used to model the developing shape of ''Arabidopsis'' leaves as they grow ([http://www.sciencemag.org/content/335/6072/1092.abstract Kuchen et al 2012]) a model that was extended to include ''Arabidopsis'' petals [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/OpenSourceDownload_PLoS_SauretGueto_2013/GPT_Petal_PLoS_20130502.zip Sauret-Güeto et al 2013]. | |||
[[Software#Quantitative understanding of growing shapes: GFtbox|<span style="color:DarkGreen;">More details on growth </span>]]<br><br> | |||
==<span style="color:DarkGreen;">Viewing three dimensional images== | |||
{| border="0" width=100% style="background-color:#000000;" | |||
|- | |||
|align="center"| | |||
[[Image:Cs0prxz0.png|32x32px]] | |||
[[Image:Leaf_trichomes.png|50px]] | |||
[[Image:Cs0prxz0.png|50px]] | |||
[[Image:GL2_GUS.png|50px]] | |||
[[Image:Leaf5.png|50px]] | |||
[[Image:OleosinSeed.png|50px]] | |||
[[Image:OPT_Leaf_copy.png|50px]] | |||
[[Image:Seedling_copy.png|50px]] | |||
[[Image:Snapdragon_Peloric_mutant.png|50px]] | |||
[[Image:Tissue.png|50px]] | |||
[[Image:Z9r3j2yx.png|50px]] | |||
[[Image:1896_wh_txr_light.png|50px]] | |||
[[Image:Ara_flower.png|50px]] | |||
[[Image:Arableaf_ath8_OPT.png|50px]] | |||
|} | |||
<br> | |||
[[Software#Viewing and measuring volume images: VolViewer|<span style="color:Green;">'''MORE'''</span>]]<br><br> | |||
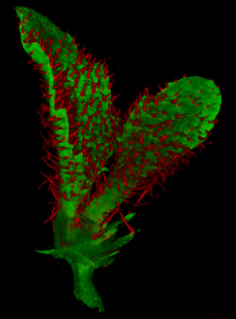
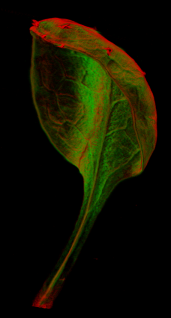
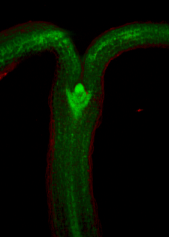
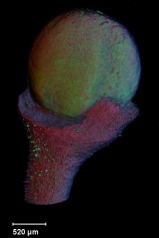
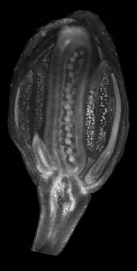
Images of plants, plant organs and cells.<br><br> | |||
''VolViewer'' uses [http://www.opengl.org/ OpenGL] and [http://qt.nokia.com/products/ Qt] to provide a user friendly application to interactively explore and quantify multi-dimensional biological images. It has been successfully used in our lab to explore and quantify confocal microscopy and optical projection tomography images. It is open-source and is compatible with the Open Microscopy Environment ([http://openmicroscopy.org/site OME]).<br><br> | |||
Movies of [http://www.youtube.com/user/InnerWorldsJIC carnivorous plants] | |||
[[Software#Viewing and measuring volume images: VolViewer|<span style="color:DarkGreen;">More details on viewing three dimensional images</span>]]<br><br> | |||
==<span style="color:DarkGreen;">Analysing shapes: faces, leaves and flowers== | |||
[[Image:PortraitsMEANSsmaller.jpg|800px]] | |||
<br> | |||
[[Software#Analysing shapes in 2D and 3D: AAMToolbox|<span style="color:Green;">'''MORE'''</span>]]<br> | |||
Seen the origional paintings? Do they exist?. <br><br> | |||
The ''AAMToolbox'' is used to analyse the shape and colour of collections of similar objects. Originally developed to analyse face shapes for lipreading ([http://ieeexplore.ieee.org/xpl/freeabs_all.jsp?arnumber=982900 Matthews ''et al''. 2002][http://www2.cmp.uea.ac.uk/~sjc/matthews-pami-01.pdf version of pdf]), we have used it extensively for analysing the shapes of leaves ([http://www.pnas.org/content/102/29/10221.short Langlade ''et al'' 2005.],[http://www.tandfonline.com/doi/abs/10.2976/1.2836738 Bensmihen ''et al.'' 2010]) and petals ([http://www.sciencemag.org/content/313/5789/963.short Whibley ''et al'' 2006],[http://www.mssaleshops.info/content/21/10/2999.short Feng ''et al''. 2010]). The analysis can be applied to art, for example, finding systematic differences between portraits by Rembrandt and Modigliani.<br><br> | |||
[[Software#Analysing shapes in 2D and 3D: AAMToolbox|<span style="color:DarkGreen;">More details on analysing shapes</span>]]<br><br> | |||
=<span style="color:Navy;">Algorithms= | |||
---- | |||
==<span style="color:Navy;">Reaction-diffusion and morphogenesis - the growth of shapes== | |||
{| border="0" width=100% style="background-color:#000000;" | |||
|- | |||
|align="center"| | |||
[[Image:tentacles_reaction_diffusion.png|400px]] | |||
[[Image:tentacles_morphogenesis.png|600px]] | |||
|} | |||
<br> | |||
[[Software#Reaction-diffusion and morphogenesis|<span style="color:Green;">'''MORE'''</span>]]<br><br> | |||
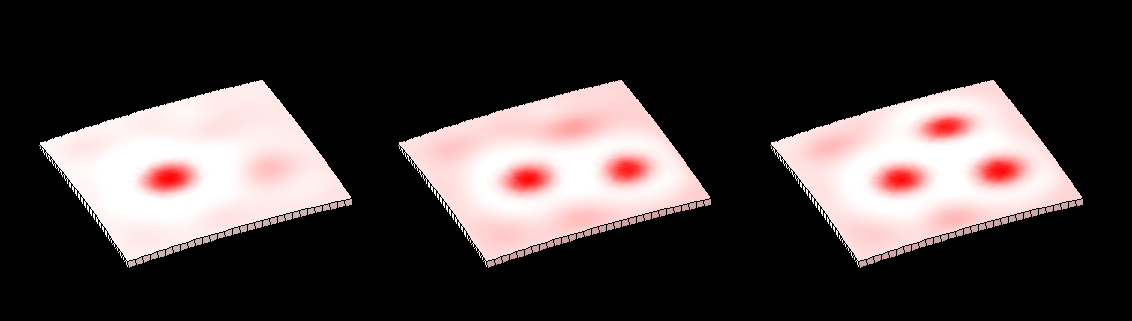
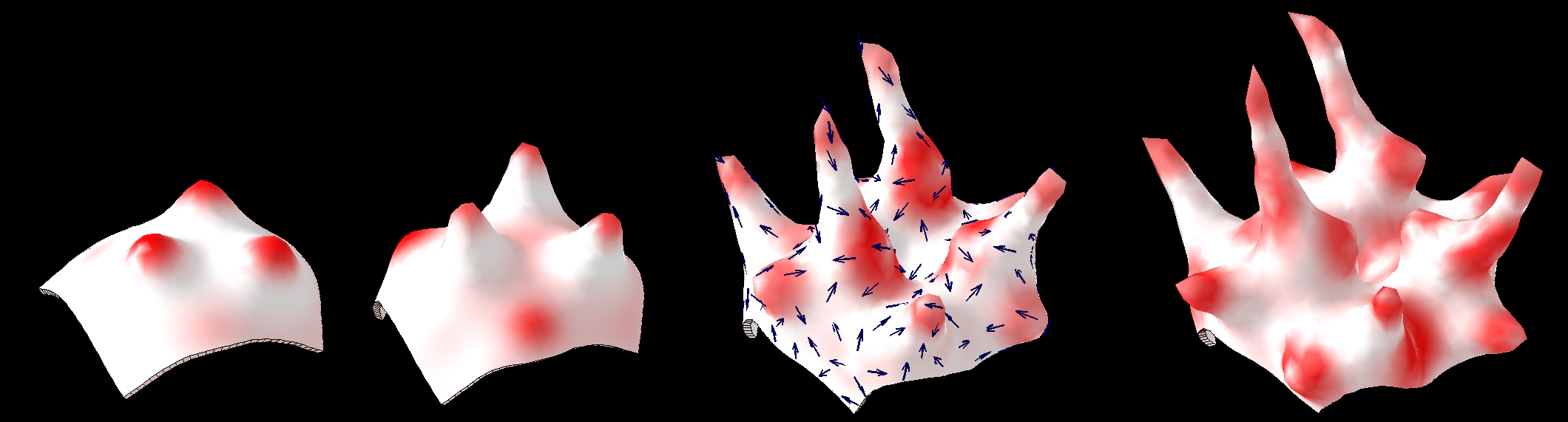
In 1952 Alan Turing proposed [http://rstb.royalsocietypublishing.org/content/237/641/37.abstract The chemical basis of Morphogenesis] - "... suggested that a system of chemical substances, called morphogens, reacting together and diffusing through a tissue, is adequate to account for the main phenomena of morphogenesis. Such a system, although it may originally be quite homogeneous, may later develop a pattern or structure due to an instability of the homogeneous equilibrium, which is triggered off by random disturbances. ..." Such patterning is now [http://en.wikipedia.org/wiki/Reaction%E2%80%93diffusion_system widely known]. However, the morphogenesis element of the story has been less widely explored - here we illustrate the process using ''GFtbox'' - but also see: [http://www.sciencedirect.com/science/article/pii/S1360138507000611 plant meristem][http://home.thep.lu.se/~henrik/mnxa09/Jonsson2012.pdf review related plant stuff] | |||
Two chemical substances react and diffuse to dynamically develop a pattern of spots (top row). We have added two simple growth rules ([http://www.ploscompbiol.org/article/info:doi/10.1371/journal.pcbi.1002071 based on our hypotheses on the growth of shapes]) to dynamically translate the pattern into a pattern of growth (bottom row). The changing geometry arising through growth which in turn feeds back on the reaction-diffusion system to modulate patterning. One of the morphogenesis rules uses the chemical concentration gradient to set the axes for anisotropic growth (arrows in third panel). | |||
This model was featured in a video interview exhibit in the London Science Museum 'Codebreakers' exhibition [http://www.sciencemuseum.org.uk/visitmuseum/galleries/turing.aspx Codebreakers]. <br> | |||
[[Software#Reaction-diffusion and morphogenesis|<span style="color:Navy;">More details on reaction-diffusion and morphogenesis</span>]]<br><br> | |||
==<span style="color:Navy;">MSER's, extrema, Connected-set filters, Sieves and '''Scale-space'''== | |||
{| border="0" width=100% style="background-color:#000000;" | |||
|- | |||
|align="center"| | |||
|} | |||
<br> | |||
In development. | |||
Revision as of 09:28, 30 September 2013
Bangham at UEA
Bangham Lab - Home
Current activity: a collaboration with the CoenLab with the aim of understanding how patterns of gene activity in biological organs influence the developing shape. The BanghamLab is focussed on the conceptual underpinning: concepts captured in computational growth models, experimental data visualisation and analysis.
Computational biology toolboxes
Growing complex biological shapes from patterns of gene expression
Movies: Model Snapdragon flower movie, Why Snapdragon flower shape is so complex
The growth of a complex snapdragon flower shape. Key to the model, is an hypothesis on how organisers control the axes along which growth occurs. The organisers are shown in cyan and green. On the right are the shapes of two symmetrical mutants computed from the same model (hypotheses).
The Growing Polarised Tissue Framework for understanding and modelling the relationship between gene activity and the growth of shapes such leaves, flowers and animal embryos is introduced in (Kennaway et al 2011). The GPT-framework was used to capture an understanding of (to model) the Snapdragon flower Green et al 2011. The Snapdragon model was validated by comparing the results with other mutant and transgenic flowers Cui et al 2010.
The GPT-framework was also used to model the developing shape of Arabidopsis leaves as they grow (Kuchen et al 2012) a model that was extended to include Arabidopsis petals Sauret-Güeto et al 2013.
Viewing three dimensional images
Images of plants, plant organs and cells.
VolViewer uses OpenGL and Qt to provide a user friendly application to interactively explore and quantify multi-dimensional biological images. It has been successfully used in our lab to explore and quantify confocal microscopy and optical projection tomography images. It is open-source and is compatible with the Open Microscopy Environment (OME).
Movies of carnivorous plants
More details on viewing three dimensional images
Analysing shapes: faces, leaves and flowers

MORE
Seen the origional paintings? Do they exist?.
The AAMToolbox is used to analyse the shape and colour of collections of similar objects. Originally developed to analyse face shapes for lipreading (Matthews et al. 2002version of pdf), we have used it extensively for analysing the shapes of leaves (Langlade et al 2005.,Bensmihen et al. 2010) and petals (Whibley et al 2006,Feng et al. 2010). The analysis can be applied to art, for example, finding systematic differences between portraits by Rembrandt and Modigliani.
More details on analysing shapes
Algorithms
Reaction-diffusion and morphogenesis - the growth of shapes
MORE
In 1952 Alan Turing proposed The chemical basis of Morphogenesis - "... suggested that a system of chemical substances, called morphogens, reacting together and diffusing through a tissue, is adequate to account for the main phenomena of morphogenesis. Such a system, although it may originally be quite homogeneous, may later develop a pattern or structure due to an instability of the homogeneous equilibrium, which is triggered off by random disturbances. ..." Such patterning is now widely known. However, the morphogenesis element of the story has been less widely explored - here we illustrate the process using GFtbox - but also see: plant meristemreview related plant stuff
Two chemical substances react and diffuse to dynamically develop a pattern of spots (top row). We have added two simple growth rules (based on our hypotheses on the growth of shapes) to dynamically translate the pattern into a pattern of growth (bottom row). The changing geometry arising through growth which in turn feeds back on the reaction-diffusion system to modulate patterning. One of the morphogenesis rules uses the chemical concentration gradient to set the axes for anisotropic growth (arrows in third panel).
This model was featured in a video interview exhibit in the London Science Museum 'Codebreakers' exhibition Codebreakers.
More details on reaction-diffusion and morphogenesis
MSER's, extrema, Connected-set filters, Sieves and Scale-space
In development.