MTtbox documentation: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 29: | Line 29: | ||
|- valign="top" | |- valign="top" | ||
|width="500pt"|A project is saved by selecting: ''menu:File:Save as''<br><br>Having first saved a project a default Interaction Function is created by selecting ''Edit''. A default project file [[MTtbox default interaction function|(example of the default interaction function)]] contains lots of comments to provide help on how to develop the project.<br><br>At present the Interaction Functions is not copied to the new project on each Save as command - this has to be done manually. | |width="500pt"|A project is saved by selecting: ''menu:File:Save as''<br><br>Having first saved a project a default Interaction Function is created by selecting ''Edit''. A default project file [[MTtbox default interaction function|(example of the default interaction function)]] contains lots of comments to provide help on how to develop the project.<br><br>At present the Interaction Functions is not copied to the new project on each Save as command - this has to be done manually. | ||
|width="400pt"|[[ | |width="400pt"|(Comments are in green - this web version of the matlab file is created using webify_interaction_function('MT_Edinb_20120427.m')).<br><br> | ||
The default file is largely a copy of''' MTtbox_BoilerPlate.txt''' which should be updated to reflect the latest ideas on how to build the function. | |||
<br><br> | |||
function data = MT_Edinb_20120427( data, before ) | |||
if nargin < 1, return; end | |||
<span style="color: Green">% </span> | |||
<span style="color: Green">%</span> | |||
<span style="color: Green">% extract factors from structure</span> | |||
for i=1:size( data.factorprops.Concentration,2) | |||
factname=lower(data.factorprops.Index2Name{i}); | |||
eval([[factname,'_p'],'=reshape(','data.factorprops.Concentration(:,i),[ data.cellprops.cellsize]);']); | |||
eval([[factname,'_i'],'=','data.factorprops.Name2Index.(factname);']); | |||
end | |||
<span style="color: Green">% insert model in here</span> | |||
<span style="color: Green">% data=setupDiffusion(data,'Factorname','s_micplm','DiffK',[0.2,0.2],...</span> | |||
<span style="color: Green">% 'RegionLists',{{'cytoplasm','vacuole','cell_wall'},{'plasma_membrane','microtubules'}});</span> | |||
<span style="color: Green">% data=setupDiffusion(data,'Factorname','id_micplm','DiffK',[0,0],...</span> | |||
<span style="color: Green">% 'RegionLists',{{'cytoplasm','vacuole','cell_wall'},{'plasma_membrane','microtubules'}});</span> | |||
<span style="color: Green">% data.factorprops.DiffusionConst(:,1)=0.1; % This should be superceed by the above</span> | |||
<span style="color: Green">% data.factorprops.DiffusionConst(:,2)=0.05;% This should be superceed by the above</span> | |||
if data.iterations<0 | |||
<span style="color: Green">% insert initialisation code here - use Initialise button to jump to here</span> | |||
<span style="color: Green">% modify default cell specifications, etc. to modify how the cell volume is partitioned, e.g.</span> | |||
<span style="color: Green">% data.organelleprops.plasma_membrane.size=0.95; % fraction of longest dimension</span> | |||
<span style="color: Green">% data.organelleprops.cytoplasm.size=0.80;</span> | |||
<span style="color: Green">% data.organelleprops.vacuole.size=0.4;</span> | |||
<span style="color: Green">% data.organelleprops.microtubules.nuclspont=0.0012; % observed nucleations per square (cubic) micron per sec</span> | |||
elseif data.iterations==0 | |||
<span style="color: Green">% insert startup code here - use Restart button to jump to here</span> | |||
<span style="color: Green">%% setup MT size</span> | |||
<span style="color: Green">%if data.cellprops.cellsize(1)>25</span> | |||
<span style="color: Green">% data=mtSETprops(data,'microtubules','size',0.01,'microtubules','growthrate',0.025);</span> | |||
<span style="color: Green">% data=mtSETprops(data,'microtubules','shrinkrate',0.005);</span> | |||
<span style="color: Green">%end</span> | |||
else | |||
<span style="color: Green">%id_micplm_p(:)=0; % clear</span> | |||
<span style="color: Green">%id_micplm_p(data.cellprops.Vol==data.organelleprops.cytoplasm.Label)=0.8; % set cytoplasm region to 1</span> | |||
<span style="color: Green">%id_micplm_p(round(data.cellprops.cellsize(1)/3):end,:,:)=0; % then set right hand end back to zero</span> | |||
<span style="color: Green">%s_micplm_p(id_micplm_p>0.1)=0.1;</span> | |||
<span style="color: Green">%% limit spontaneous nucleation to a region specified by id_micplm_p</span> | |||
<span style="color: Green">% data.organelleprops.microtubules.nuclspont=id_micplm_p;</span> | |||
<span style="color: Green">%data.cellprops.colourType=0; % set to 1 if colour is to be specified by individual MT property</span> | |||
<span style="color: Green">%Work through all the microtubules updating properties depending on flags</span> | |||
<span style="color: Green">%Average number of MT's present is observed to be about 80 i.e. 80/300 square microns (say 1 micron thick</span> | |||
<span style="color: Green">%so ~0.25 per square (cubic) micron</span> | |||
for i_MT=1:length(data.working.dyn.microtubules.Org) | |||
MT=data.working.dyn.microtubules.Org(i_MT); | |||
<span style="color: Green">% MTtbox monitors ...</span> | |||
<span style="color: Green">% MT.Props.growthrate</span> | |||
<span style="color: Green">% MT.Props.shrinkrate</span> | |||
<span style="color: Green">% MT.Props.maxbend</span> | |||
<span style="color: Green">% MT.Props.maxAge</span> | |||
<span style="color: Green">% MT.Props.nuclspont</span> | |||
<span style="color: Green">% MT.Props.nuclonMT</span> | |||
<span style="color: Green">% Make changes here, i.e.</span> | |||
<span style="color: Green">% diffs=diff(MT.Verts).^2; % calculate length of MT</span> | |||
<span style="color: Green">% MTlength=sum(sqrt(sum(diffs')'))*data.cellprops.micronsPerVoxelEdge; % microns</span> | |||
<span style="color: Green">% if MTlength>0.5</span> | |||
<span style="color: Green">% MT.Props.nuclonMT=min(1,0.4*MTlength); % observed approx. 0.007 per micron per sec</span> | |||
<span style="color: Green">% else</span> | |||
<span style="color: Green">% MT.Props.nuclonMT=0;</span> | |||
<span style="color: Green">% end</span> | |||
<span style="color: Green">%ageMT=data.working.CellAge;% seconds</span> | |||
<span style="color: Green">%maxAgeColour=20;</span> | |||
<span style="color: Green">%MT.Props.FaceColor=[max(0,min(1,1-ageMT/maxAgeColour)), 0.5, max(0,min(1,ageMT/maxAgeColour))];</span> | |||
<span style="color: Green">% if MT.BoundPLM %&& MT.Growing % it must have hit the plasma_membrane and not aligned</span> | |||
<span style="color: Green">% MT.Props.shrinkrate=20; % force it to shrink to oblivion</span> | |||
<span style="color: Green">% fprintf(1,'MT(%d) Collided with PLM fastshrink\n',i_MT);</span> | |||
<span style="color: Green">% end</span> | |||
<span style="color: Green">% if MT.BoundMic %&& MT.Growing % it must have hit the plasma_membrane and not aligned</span> | |||
<span style="color: Green">% MT.Props.shrinkrate=20; % force it to shrink to oblivion</span> | |||
<span style="color: Green">% fprintf(1,'MT(%d) Collided with MT fastshrink\n',i_MT);</span> | |||
<span style="color: Green">% end</span> | |||
<span style="color: Green">% Keep the following</span> | |||
<span style="color: Green">% Clear flags from collision detector</span> | |||
MT.BoundPLM=false; | |||
MT.BoundMic=false; | |||
MT.BoundVac=false; | |||
data.working.dyn.microtubules.Org(i_MT)=MT; | |||
end | |||
end | |||
<span style="color: Green">% put factors back into structure</span> | |||
for i=1:size( data.factorprops.Concentration,2) | |||
factname=lower(data.factorprops.Index2Name{i}); | |||
eval(['data.factorprops.Concentration(:,i)=',[factname,'_p(:);']]); | |||
end | |||
end | |||
<span style="color: Green">% This space intentionally left blank.</span> | |||
<span style="color: Green">% Default factor parameters</span> | |||
<span style="color: Green">% Default organelle parameters</span> | |||
<span style="color: Green">% cell_wall.size =0.000000</span> | |||
<span style="color: Green">% cell_wall.offset =0.500000</span> | |||
<span style="color: Green">% cell_wall.micronthick =0.100000</span> | |||
<span style="color: Green">% cell_wall.minvox =1.000000</span> | |||
<span style="color: Green">% cell_wall.FaceColor =[1.000000,0.000000,1.000000,]</span> | |||
<span style="color: Green">% cell_wall.FaceAlpha =0.010000</span> | |||
<span style="color: Green">% cell_wall.Label =-5.000000</span> | |||
<span style="color: Green">% cell_wall.isLayer =1.000000</span> | |||
<span style="color: Green">% cell_wall.Static =1.000000</span> | |||
<span style="color: Green">% cell_wall.InUse =1.000000</span> | |||
<span style="color: Green">%plasma_membrane.size =0.000000</span> | |||
<span style="color: Green">%plasma_membrane.offset =0.500000</span> | |||
<span style="color: Green">%plasma_membrane.micronthick =0.010000</span> | |||
<span style="color: Green">%plasma_membrane.minvox =3.000000</span> | |||
<span style="color: Green">%plasma_membrane.FaceColor =[1.000000,1.000000,0.000000,]</span> | |||
<span style="color: Green">%plasma_membrane.FaceAlpha =0.020000</span> | |||
<span style="color: Green">%plasma_membrane.Label =-4.000000</span> | |||
<span style="color: Green">%plasma_membrane.isLayer =1.000000</span> | |||
<span style="color: Green">%plasma_membrane.Static =1.000000</span> | |||
<span style="color: Green">%plasma_membrane.InUse =1.000000</span> | |||
<span style="color: Green">% cytoplasm.size =0.000000</span> | |||
<span style="color: Green">% cytoplasm.offset =0.500000</span> | |||
<span style="color: Green">% cytoplasm.micronthick =1.000000</span> | |||
<span style="color: Green">% cytoplasm.minvox =5.000000</span> | |||
<span style="color: Green">% cytoplasm.FaceColor =[0.000000,1.000000,0.000000,]</span> | |||
<span style="color: Green">% cytoplasm.FaceAlpha =0.020000</span> | |||
<span style="color: Green">% cytoplasm.Label =0.000000</span> | |||
<span style="color: Green">% cytoplasm.isLayer =1.000000</span> | |||
<span style="color: Green">% cytoplasm.Static =1.000000</span> | |||
<span style="color: Green">% cytoplasm.InUse =1.000000</span> | |||
<span style="color: Green">% microtubules.size =0.025000</span> | |||
<span style="color: Green">% microtubules.minvoxel =3.000000</span> | |||
<span style="color: Green">% microtubules.growthrate =0.100000</span> | |||
<span style="color: Green">% microtubules.shrinkrate =0.010000</span> | |||
<span style="color: Green">% microtubules.maxbend =40.000000</span> | |||
<span style="color: Green">% microtubules.MaxAngle =20.000000</span> | |||
<span style="color: Green">% microtubules.maxAge =0.000000</span> | |||
<span style="color: Green">% microtubules.nuclspont =0.050000</span> | |||
<span style="color: Green">% microtubules.nuclonMT =0.090000</span> | |||
<span style="color: Green">% microtubules.probCatastrophe=0.300000</span> | |||
<span style="color: Green">% microtubules.FaceColor =[0.000000,0.000000,1.000000,]</span> | |||
<span style="color: Green">% microtubules.FaceAlpha =0.400000</span> | |||
<span style="color: Green">% microtubules.Label =-6.000000</span> | |||
<span style="color: Green">% microtubules.isLayer =0.000000</span> | |||
<span style="color: Green">% microtubules.Static =0.000000</span> | |||
<span style="color: Green">% microtubules.InUse =1.000000</span> | |||
<span style="color: Green">% Default cell parameters</span> | |||
<span style="color: Green">% cellprops.shape ='Sphere'</span> | |||
<span style="color: Green">% cellprops.sheetmodel =0</span> | |||
<span style="color: Green">% cellprops.maxLengthMicrons=20</span> | |||
<span style="color: Green">% cellprops.secondsPerStep =1</span> | |||
<span style="color: Green">% cellprops.micronsPerVoxelEdge=0.800000</span> | |||
<span style="color: Green">% cellprops.cubicMicronsPerVoxel=0.512000</span> | |||
<span style="color: Green">% cellprops.cellsize =[25,25,25,]</span> | |||
<span style="color: Green">% cellprops.colourType =1</span> | |||
<span style="color: Green">% cellprops.Vol =[-1,-1,-1,-1, ... ]</span> | |||
<span style="color: Green">% cellprops.Smooth ='None'</span> | |||
<span style="color: Green">% cellprops.dynamic ='microtubules'...</span> | |||
<span style="color: Green">% cellprops.bound_distance =5</span> | |||
<span style="color: Green">% cellprops.collide_distance=10</span> | |||
<span style="color: Green">% cellprops.% vacuole.Vol=struct</span> | |||
<span style="color: Green">% vacuole.EdgeVol=struct</span> | |||
<span style="color: Green">% cellprops.% cell_wall.Vol=struct</span> | |||
<span style="color: Green">% cell_wall.EdgeVol=struct</span> | |||
<span style="color: Green">% cellprops.% plasma_membrane.Vol=struct</span> | |||
<span style="color: Green">% plasma_membrane.EdgeVol=struct</span> | |||
<span style="color: Green">% cellprops.% cytoplasm.Vol=struct</span> | |||
<span style="color: Green">% cytoplasm.EdgeVol=struct</span> | |||
<span style="color: Green">% cytoplasm.Indexes=struct</span> | |||
<span style="color: Green">% cellprops.% Diameter.microtubules=struct</span> | |||
<span style="color: Green">% cellprops.% microtubules.Vol=struct</span> | |||
<span style="color: Green">% microtubules.Offsets=struct</span> | |||
<span style="color: Green">% cellprops.sheetPlaneNormal=[0,1,0,]</span> | |||
|} | |} | ||
Revision as of 16:15, 27 April 2012
Return to Bangham Lab Software
Current Status
MTtbox is currently under test and further development
The main data structure is called: 'data'. It can be accessed from the Matlab command line by declaring data to be global.
global data
at any time. The following documentation will refer to fields in data. It also refers to the custom menu items by menu:name.
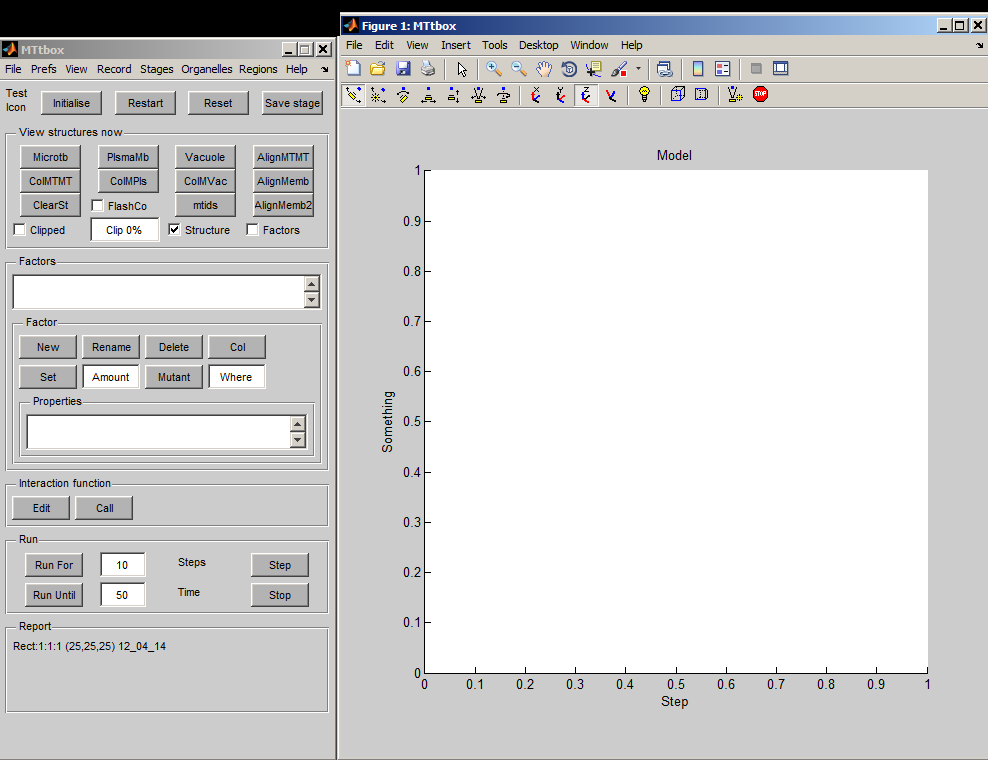
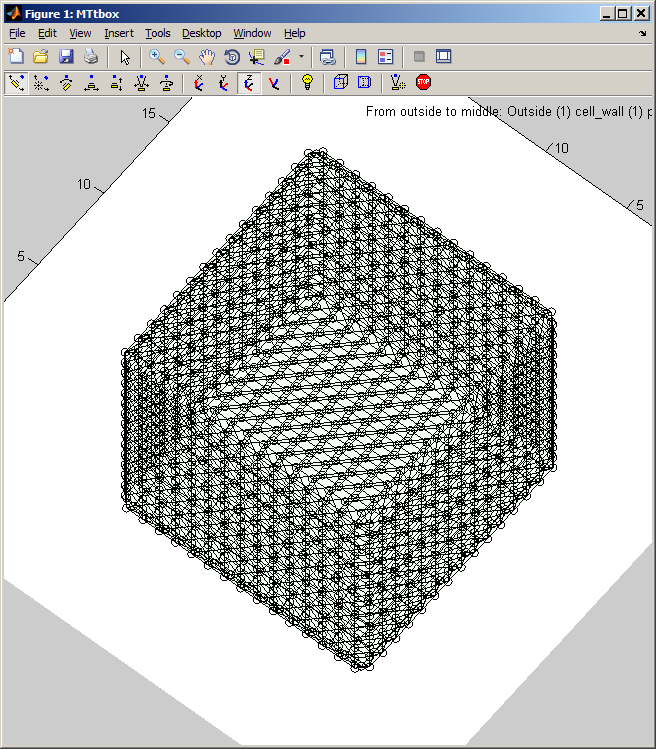
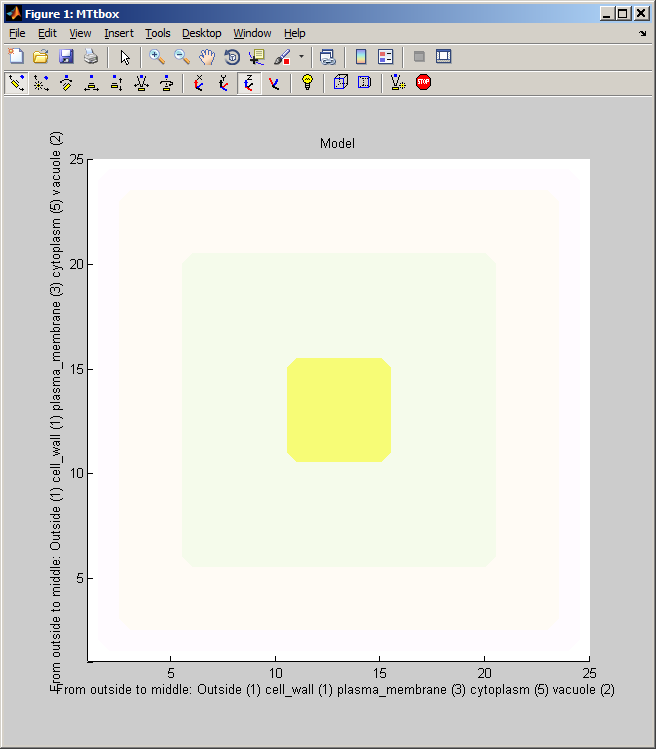
Graphical User Interface
1 A
1 B
1 C
| A project is saved by selecting: menu:File:Save as Having first saved a project a default Interaction Function is created by selecting Edit. A default project file (example of the default interaction function) contains lots of comments to provide help on how to develop the project. At present the Interaction Functions is not copied to the new project on each Save as command - this has to be done manually. |
(Comments are in green - this web version of the matlab file is created using webify_interaction_function('MT_Edinb_20120427.m')). The default file is largely a copy of MTtbox_BoilerPlate.txt which should be updated to reflect the latest ideas on how to build the function.
function data = MT_Edinb_20120427( data, before )
if nargin < 1, return; end
%
%
% extract factors from structure
for i=1:size( data.factorprops.Concentration,2)
factname=lower(data.factorprops.Index2Name{i});
eval([[factname,'_p'],'=reshape(','data.factorprops.Concentration(:,i),[ data.cellprops.cellsize]);']);
eval([[factname,'_i'],'=','data.factorprops.Name2Index.(factname);']);
end
% insert model in here
% data=setupDiffusion(data,'Factorname','s_micplm','DiffK',[0.2,0.2],...
% 'RegionLists',{{'cytoplasm','vacuole','cell_wall'},{'plasma_membrane','microtubules'}});
% data=setupDiffusion(data,'Factorname','id_micplm','DiffK',[0,0],...
% 'RegionLists',{{'cytoplasm','vacuole','cell_wall'},{'plasma_membrane','microtubules'}});
% data.factorprops.DiffusionConst(:,1)=0.1; % This should be superceed by the above
% data.factorprops.DiffusionConst(:,2)=0.05;% This should be superceed by the above
if data.iterations<0
% insert initialisation code here - use Initialise button to jump to here
% modify default cell specifications, etc. to modify how the cell volume is partitioned, e.g.
% data.organelleprops.plasma_membrane.size=0.95; % fraction of longest dimension
% data.organelleprops.cytoplasm.size=0.80;
% data.organelleprops.vacuole.size=0.4;
% data.organelleprops.microtubules.nuclspont=0.0012; % observed nucleations per square (cubic) micron per sec
elseif data.iterations==0
% insert startup code here - use Restart button to jump to here
%% setup MT size
%if data.cellprops.cellsize(1)>25
% data=mtSETprops(data,'microtubules','size',0.01,'microtubules','growthrate',0.025);
% data=mtSETprops(data,'microtubules','shrinkrate',0.005);
%end
else
%id_micplm_p(:)=0; % clear
%id_micplm_p(data.cellprops.Vol==data.organelleprops.cytoplasm.Label)=0.8; % set cytoplasm region to 1
%id_micplm_p(round(data.cellprops.cellsize(1)/3):end,:,:)=0; % then set right hand end back to zero
%s_micplm_p(id_micplm_p>0.1)=0.1;
%% limit spontaneous nucleation to a region specified by id_micplm_p
% data.organelleprops.microtubules.nuclspont=id_micplm_p;
%data.cellprops.colourType=0; % set to 1 if colour is to be specified by individual MT property
%Work through all the microtubules updating properties depending on flags
%Average number of MT's present is observed to be about 80 i.e. 80/300 square microns (say 1 micron thick
%so ~0.25 per square (cubic) micron
for i_MT=1:length(data.working.dyn.microtubules.Org)
MT=data.working.dyn.microtubules.Org(i_MT);
% MTtbox monitors ...
% MT.Props.growthrate
% MT.Props.shrinkrate
% MT.Props.maxbend
% MT.Props.maxAge
% MT.Props.nuclspont
% MT.Props.nuclonMT
% Make changes here, i.e.
% diffs=diff(MT.Verts).^2; % calculate length of MT
% MTlength=sum(sqrt(sum(diffs')'))*data.cellprops.micronsPerVoxelEdge; % microns
% if MTlength>0.5
% MT.Props.nuclonMT=min(1,0.4*MTlength); % observed approx. 0.007 per micron per sec
% else
% MT.Props.nuclonMT=0;
% end
%ageMT=data.working.CellAge;% seconds
%maxAgeColour=20;
%MT.Props.FaceColor=[max(0,min(1,1-ageMT/maxAgeColour)), 0.5, max(0,min(1,ageMT/maxAgeColour))];
% if MT.BoundPLM %&& MT.Growing % it must have hit the plasma_membrane and not aligned
% MT.Props.shrinkrate=20; % force it to shrink to oblivion
% fprintf(1,'MT(%d) Collided with PLM fastshrink\n',i_MT);
% end
% if MT.BoundMic %&& MT.Growing % it must have hit the plasma_membrane and not aligned
% MT.Props.shrinkrate=20; % force it to shrink to oblivion
% fprintf(1,'MT(%d) Collided with MT fastshrink\n',i_MT);
% end
% Keep the following
% Clear flags from collision detector
MT.BoundPLM=false;
MT.BoundMic=false;
MT.BoundVac=false;
data.working.dyn.microtubules.Org(i_MT)=MT;
end
end
% put factors back into structure
for i=1:size( data.factorprops.Concentration,2)
factname=lower(data.factorprops.Index2Name{i});
eval(['data.factorprops.Concentration(:,i)=',[factname,'_p(:);']]);
end
end
% This space intentionally left blank.
% Default factor parameters
% Default organelle parameters
% cell_wall.size =0.000000
% cell_wall.offset =0.500000
% cell_wall.micronthick =0.100000
% cell_wall.minvox =1.000000
% cell_wall.FaceColor =[1.000000,0.000000,1.000000,]
% cell_wall.FaceAlpha =0.010000
% cell_wall.Label =-5.000000
% cell_wall.isLayer =1.000000
% cell_wall.Static =1.000000
% cell_wall.InUse =1.000000
%plasma_membrane.size =0.000000
%plasma_membrane.offset =0.500000
%plasma_membrane.micronthick =0.010000
%plasma_membrane.minvox =3.000000
%plasma_membrane.FaceColor =[1.000000,1.000000,0.000000,]
%plasma_membrane.FaceAlpha =0.020000
%plasma_membrane.Label =-4.000000
%plasma_membrane.isLayer =1.000000
%plasma_membrane.Static =1.000000
%plasma_membrane.InUse =1.000000
% cytoplasm.size =0.000000
% cytoplasm.offset =0.500000
% cytoplasm.micronthick =1.000000
% cytoplasm.minvox =5.000000
% cytoplasm.FaceColor =[0.000000,1.000000,0.000000,]
% cytoplasm.FaceAlpha =0.020000
% cytoplasm.Label =0.000000
% cytoplasm.isLayer =1.000000
% cytoplasm.Static =1.000000
% cytoplasm.InUse =1.000000
% microtubules.size =0.025000
% microtubules.minvoxel =3.000000
% microtubules.growthrate =0.100000
% microtubules.shrinkrate =0.010000
% microtubules.maxbend =40.000000
% microtubules.MaxAngle =20.000000
% microtubules.maxAge =0.000000
% microtubules.nuclspont =0.050000
% microtubules.nuclonMT =0.090000
% microtubules.probCatastrophe=0.300000
% microtubules.FaceColor =[0.000000,0.000000,1.000000,]
% microtubules.FaceAlpha =0.400000
% microtubules.Label =-6.000000
% microtubules.isLayer =0.000000
% microtubules.Static =0.000000
% microtubules.InUse =1.000000
% Default cell parameters
% cellprops.shape ='Sphere'
% cellprops.sheetmodel =0
% cellprops.maxLengthMicrons=20
% cellprops.secondsPerStep =1
% cellprops.micronsPerVoxelEdge=0.800000
% cellprops.cubicMicronsPerVoxel=0.512000
% cellprops.cellsize =[25,25,25,]
% cellprops.colourType =1
% cellprops.Vol =[-1,-1,-1,-1, ... ]
% cellprops.Smooth ='None'
% cellprops.dynamic ='microtubules'...
% cellprops.bound_distance =5
% cellprops.collide_distance=10
% cellprops.% vacuole.Vol=struct
% vacuole.EdgeVol=struct
% cellprops.% cell_wall.Vol=struct
% cell_wall.EdgeVol=struct
% cellprops.% plasma_membrane.Vol=struct
% plasma_membrane.EdgeVol=struct
% cellprops.% cytoplasm.Vol=struct
% cytoplasm.EdgeVol=struct
% cytoplasm.Indexes=struct
% cellprops.% Diameter.microtubules=struct
% cellprops.% microtubules.Vol=struct
% microtubules.Offsets=struct
% cellprops.sheetPlaneNormal=[0,1,0,]
|