|
|
| (50 intermediate revisions by 2 users not shown) |
| Line 1: |
Line 1: |
| =[http://www.uea.ac.uk/computing/People/Honorary/Andrew+Bangham Bangham at UEA]=
| |
| =<span style="color:Indigo;">Bangham Lab - Home=
| |
|
| |
|
| <span style="color: DarkGreen">'''Current activity: a collaboration''' with the [http://rico-coen.jic.ac.uk/index.php/Main_Page CoenLab] with the aim of understanding how patterns of gene activity in biological organs influence the developing shape. The BanghamLab is focussed on the conceptual underpinning: concepts captured in computational growth models, experimental data visualisation and analysis.</span>
| | |
| =<span style="color:DarkGreen;">Computational biology toolboxes= | | =<span style="color:DarkGreen;">Computational biology</span>= |
|
| |
|
| ---- | | ---- |
|
| |
|
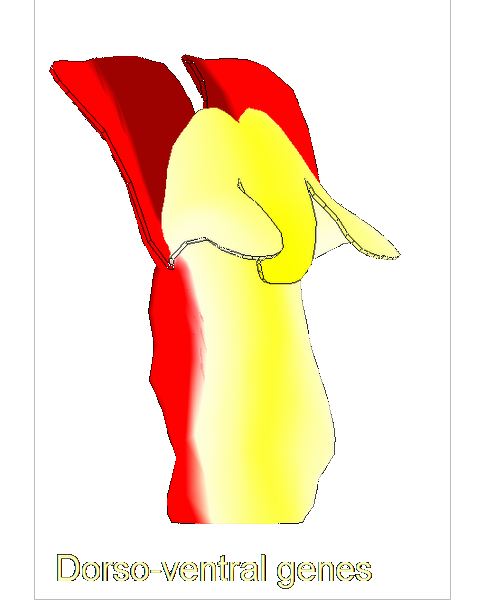
| ==<span style="color:DarkGreen;">Growing complex biological shapes from patterns of gene expression</span>== | | ==<span style="color:DarkGreen;">[[Software#Quantitative understanding of growing shapes: GFtbox|<span style="color:Green;"> '''Growing''']] complex biological shapes from patterns of gene expression</span>== |
| {| border="0" width=100% style="background-color:#000000;" | | {| border="0" width=100% style="background-color:#000000;" |
| |- | | |- |
| Line 21: |
Line 19: |
| [[Software#Quantitative understanding of growing shapes: GFtbox|<span style="color:Green;">'''MORE'''</span>]]<br> | | [[Software#Quantitative understanding of growing shapes: GFtbox|<span style="color:Green;">'''MORE'''</span>]]<br> |
|
| |
|
| Movies: [http://www.youtube.com/watch?v=7Uu5dHxyEwk Model Snapdragon flower movie], [http://www.youtube.com/watch?feature=endscreen&v=kZ45R1UlohM&NR=1 Why Snapdragon flower shape is so complex]<br><br>
| | ==<span style="color:DarkGreen;">[[Software#Viewing and measuring volume images: VolViewer|<span style="color:Green;"> '''Viewing''']] three dimensional volume (microscopy) images== |
| | |
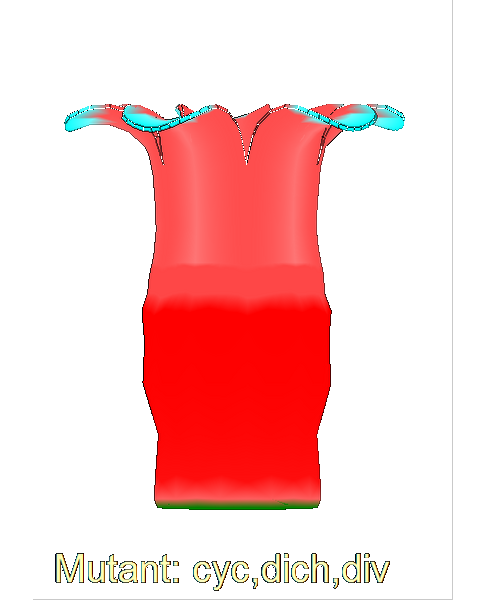
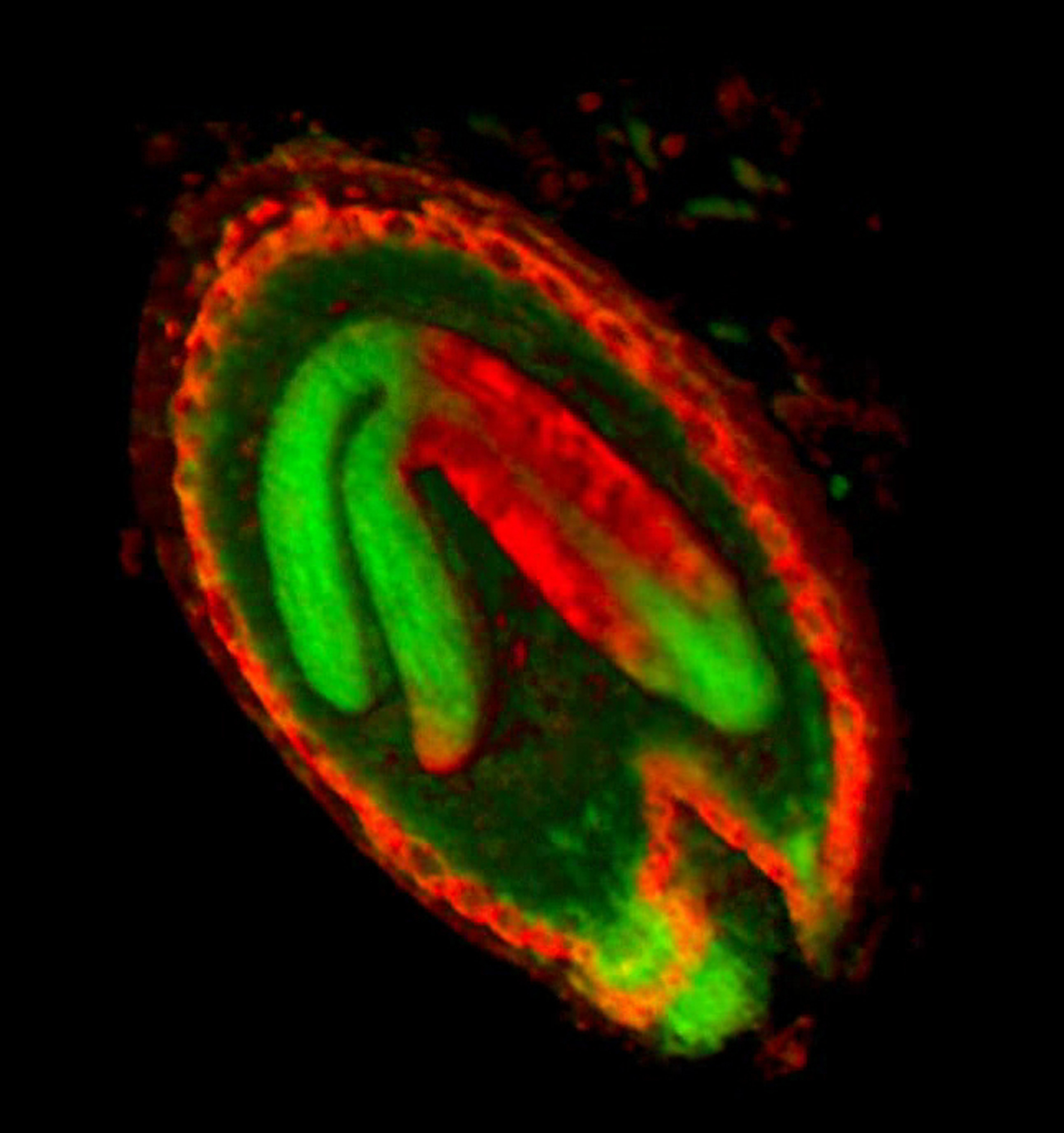
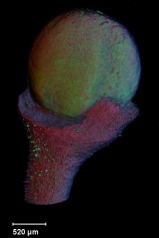
| The growth of a complex ''snapdragon flower shape''. Key to the model, is an hypothesis on'' how organisers control the axes'' along which growth occurs. The organisers are shown in cyan and green. On the right are the shapes of two symmetrical mutants computed from the same model (hypotheses).<br><br>
| |
| The Growing Polarised Tissue Framework for understanding and modelling the relationship between gene activity and the growth of shapes such leaves, flowers and animal embryos is introduced in ([http://www.ploscompbiol.org/article/info:doi/10.1371/journal.pcbi.1002071 Kennaway et al 2011]). The GPT-framework was used to capture an understanding of (to model) the Snapdragon flower [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1000537 Green et al 2011]. The Snapdragon model was validated by comparing the results with other mutant and transgenic flowers [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1000538 Cui et al 2010.]<br><br>
| |
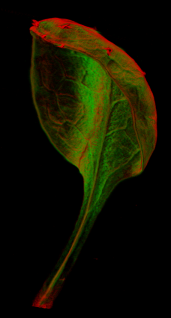
| The GPT-framework was also used to model the developing shape of ''Arabidopsis'' leaves as they grow ([http://www.sciencemag.org/content/335/6072/1092.abstract Kuchen et al 2012]) a model that was extended to include ''Arabidopsis'' petals [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/OpenSourceDownload_PLoS_SauretGueto_2013/GPT_Petal_PLoS_20130502.zip Sauret-Güeto et al 2013].
| |
| | |
| [[Software#Quantitative understanding of growing shapes: GFtbox|<span style="color:DarkGreen;">More details on growth </span>]]<br><br>
| |
| ==<span style="color:DarkGreen;">Viewing three dimensional images==
| |
| {| border="0" width=100% style="background-color:#000000;" | | {| border="0" width=100% style="background-color:#000000;" |
| |- | | |- |
| Line 48: |
Line 39: |
| |} | | |} |
| <br> | | <br> |
| [[Software#Viewing and measuring volume images: VolViewer|<span style="color:Green;">'''MORE'''</span>]]<br><br> | | [[Software#Viewing and measuring volume images: VolViewer|<span style="color:Green;">'''MORE'''</span>]] |
|
| |
|
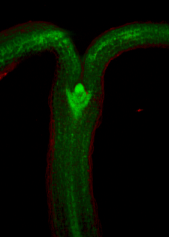
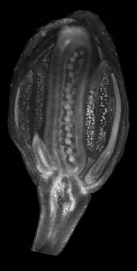
| Images of plants, plant organs and cells.<br><br>
| | ==[[Software#Analysing shapes in 2D and 3D: AAMToolbox|<span style="color:Green;">'''Analysing'''</span>]] shapes: faces, leaves and flowers== |
| ''VolViewer'' uses [http://www.opengl.org/ OpenGL] and [http://qt.nokia.com/products/ Qt] to provide a user friendly application to interactively explore and quantify multi-dimensional biological images. It has been successfully used in our lab to explore and quantify confocal microscopy and optical projection tomography images. It is open-source and is compatible with the Open Microscopy Environment ([http://openmicroscopy.org/site OME]).<br><br>
| | {| border="0" width=100% style="background-color:#000000;" |
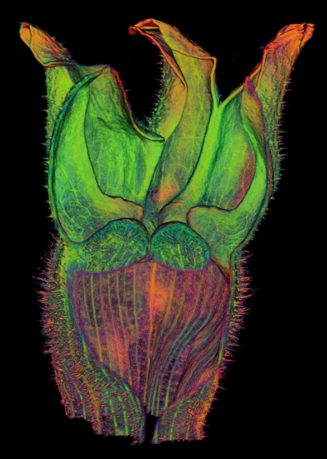
| Movies of [http://www.youtube.com/user/InnerWorldsJIC carnivorous plants]
| | |- |
| | | [[Image:PortraitsMEANSsmaller.jpg|800px]] |
| [[Software#Viewing and measuring volume images: VolViewer|<span style="color:DarkGreen;">More details on viewing three dimensional images</span>]]<br><br> | | |-} |
| | |
| ==<span style="color:DarkGreen;">Analysing shapes: faces, leaves and flowers== | |
| [[Image:PortraitsMEANSsmaller.jpg|800px]] | |
| <br> | | <br> |
| [[Software#Analysing shapes in 2D and 3D: AAMToolbox|<span style="color:Green;">'''MORE'''</span>]]<br> | | [[Software#Analysing shapes in 2D and 3D: AAMToolbox|<span style="color:Green;">'''MORE'''</span>]]<br> |
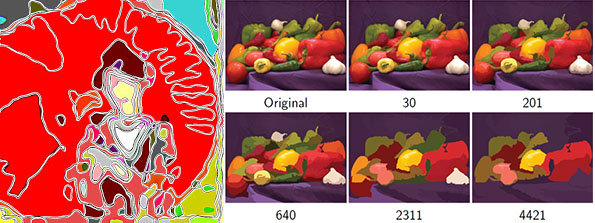
| Seen the origional paintings? Do they exist?. <br><br>
| | Have you seen the original paintings? Do they exist?. <br><br> |
| | |
| The ''AAMToolbox'' is used to analyse the shape and colour of collections of similar objects. Originally developed to analyse face shapes for lipreading ([http://ieeexplore.ieee.org/xpl/freeabs_all.jsp?arnumber=982900 Matthews ''et al''. 2002][http://www2.cmp.uea.ac.uk/~sjc/matthews-pami-01.pdf version of pdf]), we have used it extensively for analysing the shapes of leaves ([http://www.pnas.org/content/102/29/10221.short Langlade ''et al'' 2005.],[http://www.tandfonline.com/doi/abs/10.2976/1.2836738 Bensmihen ''et al.'' 2010]) and petals ([http://www.sciencemag.org/content/313/5789/963.short Whibley ''et al'' 2006],[http://www.mssaleshops.info/content/21/10/2999.short Feng ''et al''. 2010]). The analysis can be applied to art, for example, finding systematic differences between portraits by Rembrandt and Modigliani.<br><br>
| |
| [[Software#Analysing shapes in 2D and 3D: AAMToolbox|<span style="color:DarkGreen;">More details on analysing shapes</span>]]<br><br>
| |
|
| |
|
| =<span style="color:Navy;">Algorithms= | | =<span style="color:Navy;">Algorithms= |
|
| |
|
| ---- | | ---- |
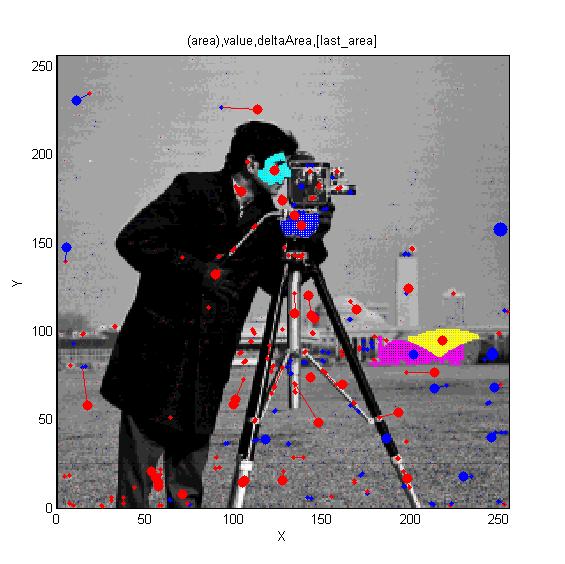
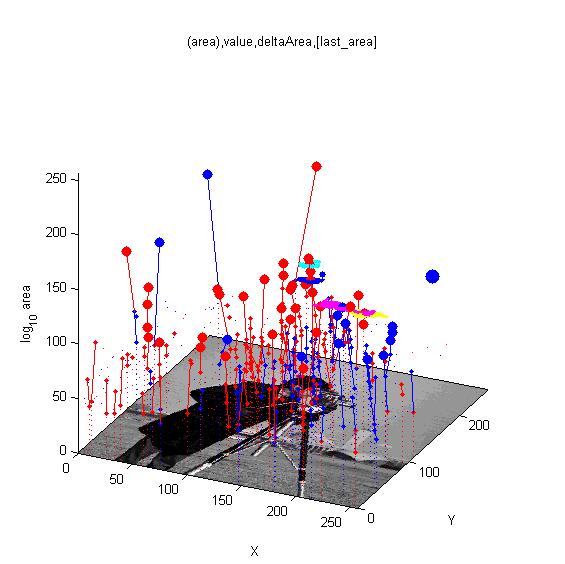
| ==<span style="color:Navy;">Reaction-diffusion and morphogenesis - the growth of shapes== | | ==[http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software#MSERs.2C_extrema.2C_connected-set_filters_and_sieves <span style="color:Navy;">'''Vision''':] MSER's, extrema, filter-banks, Sieves and '''Scale-space'''== |
| {| border="0" width=100% style="background-color:#000000;" | | {| border="0" width=100% style="background-color:#ffffff;" |
| |- | | |- |
| |align="center"| | | |align="center"| |
| [[Image:tentacles_reaction_diffusion.png|400px]] | | [[Image:Cameraman_iso_topview.jpg|300px|AAMToolbox]] |
| [[Image:tentacles_morphogenesis.png|600px]] | | [[Image:Cameraman_iso_tree.jpg|300px|AAMToolbox]] |
| |} | | |} |
| <br>
| |
| [[Software#Reaction-diffusion and morphogenesis|<span style="color:Green;">'''MORE'''</span>]]<br><br>
| |
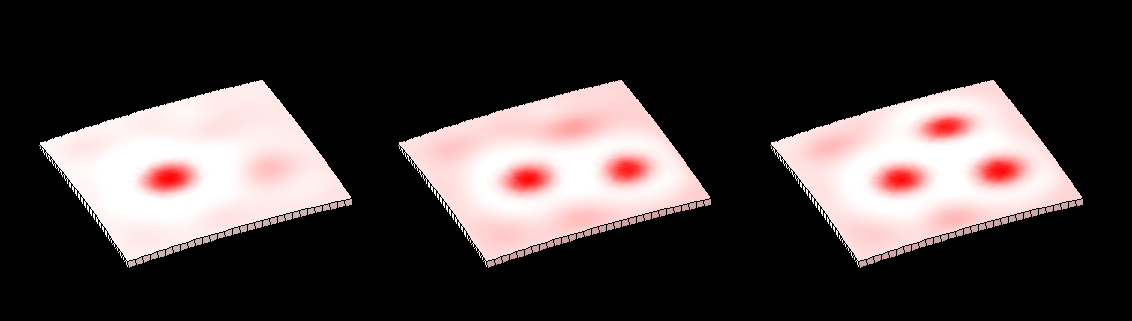
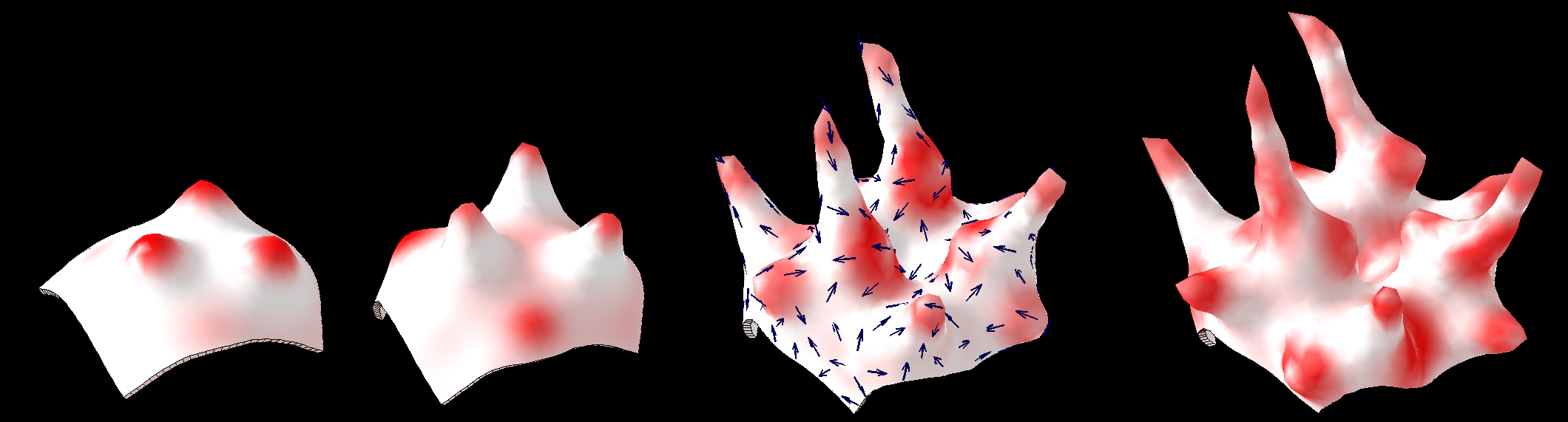
| In 1952 Alan Turing proposed [http://rstb.royalsocietypublishing.org/content/237/641/37.abstract The chemical basis of Morphogenesis] - "... suggested that a system of chemical substances, called morphogens, reacting together and diffusing through a tissue, is adequate to account for the main phenomena of morphogenesis. Such a system, although it may originally be quite homogeneous, may later develop a pattern or structure due to an instability of the homogeneous equilibrium, which is triggered off by random disturbances. ..." Such patterning is now [http://en.wikipedia.org/wiki/Reaction%E2%80%93diffusion_system widely known]. However, the morphogenesis element of the story has been less widely explored - here we illustrate the process using ''GFtbox'' - but also see: [http://www.sciencedirect.com/science/article/pii/S1360138507000611 plant meristem][http://home.thep.lu.se/~henrik/mnxa09/Jonsson2012.pdf review related plant stuff]
| |
|
| |
|
| Two chemical substances react and diffuse to dynamically develop a pattern of spots (top row). We have added two simple growth rules ([http://www.ploscompbiol.org/article/info:doi/10.1371/journal.pcbi.1002071 based on our hypotheses on the growth of shapes]) to dynamically translate the pattern into a pattern of growth (bottom row). The changing geometry arising through growth which in turn feeds back on the reaction-diffusion system to modulate patterning. One of the morphogenesis rules uses the chemical concentration gradient to set the axes for anisotropic growth (arrows in third panel).
| | [[Software#MSERs.2C_extrema.2C_connected-set_filters_and_sieves|<span style="color:Navy;">'''MORE'''</span>]] |
|
| |
|
| This model was featured in a video interview exhibit in the London Science Museum 'Codebreakers' exhibition [http://www.sciencemuseum.org.uk/visitmuseum/galleries/turing.aspx Codebreakers]. <br>
| | ==[http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software#Art.2C_extrema_of_light_and_shade:_PhotoArtMaster <span style="color:Navy;">'''Applications'''</span>]' <span style="color:Navy;">of non-linear filter banks (sieves) and the art of light and shade</span>== |
| [[Software#Reaction-diffusion and morphogenesis|<span style="color:Navy;">More details on reaction-diffusion and morphogenesis</span>]]<br><br>
| | {| border="0" width=100% style="background-color:#ffffff;" |
| ==<span style="color:Navy;">MSER's, extrema, Connected-set filters, Sieves and '''Scale-space'''==
| |
| {| border="0" width=100% style="background-color:#000000;" | |
| |- | | |- |
| |align="center"| | | |align="center"| |
| | | [[Image:Colour_sieve.jpg|600px|AAMToolbox]] |
| |} | | |} |
| <br>
| | These images were produced from photographs using '''ArtMaster''' (formally known as '''PhotoArtMaster'''). The software received many favourable reviews when it was released (e.g. [http://graphicssoft.about.com/cs/photoart/gr/photoartmasterg.htm "This software can give you a lot of satisfaction from your everyday photos"], [http://graphicssoft.about.com/library/products/aafpr_photoartmaster1.htm] |
| [[Software#MSERs extrema connected-set filters and sieves|<span style="color:Green;">'''MORE'''</span>]]<br><br>
| |
| (This is work we did a while ago, a ''blast from the past''. I failed to popularise it at the time, however, '''MSER's are now attracting lots of attention''' so I'm now contributing my bit here in a hurry - [[http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Andrews_Organ_Recital Why the hurry?]]<br><br> | |
| The algorithm for finding Maximally stable extremal regions (MSER's) '''is the same as an 'o' sieve'''. The terminology is confused. Such algorithms relate closely to mathematical morphology (openings, closings and in particular watersheds and reconstruction filters). In mathematical morphology the MSER algorithm ('o' sieve) might be called a 'connected-set opening'. It is one of a family of closely related algorithms which for which I coined the term '''sieves'''. Why? I was trying to unhook people from a misconception about 'filters'.<br><br>
| |
|
| |
|
| At the time (and now) some people assert that Gaussian filters are '''unique''': the only scale-space preserving filters. So then it was '''nice to find''' that brains might also have Gaussian filter banks - computer scientists and biologists had a comfortable agreement that it had to be so. 'Uniqueness of the gaussian kernel for scale-space filtering' is a strong title. The word ''linear'' is missing. Sieves are non-linear and '''all sieves are also scale-space preserving'''. This is one of the properties that contributes to their success as feature finders.<br><br>
| | [http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software#The_final_version_of_the_Windows_ArtMaster2.0_is_downloadable_here_with_no_support The final (so far unpublished) version of ArtMaster including code is downloadable from here.] I cannot provide support but quite of lot of documentation is available within [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/SieveWebPages/a4a_2_screensize.pdf <span style="color: Chocolate">''''this document''''' </span>] |
|
| |
|
| 2D MSER's (sieves in our old terminology) are used for finding objects in. Sieves have also been used, e.g. in 1D: for analysing ID protein hydrophobicity plots(Bangham, 1988<ref>Bangham, J.A. (1988). ''Data-sieving hydrophobicity plots. Anal. Biochem''. 174, 142–145</ref>), de-noising single channel current data(Bangham et al, 1984<ref>Bangham, J.A., and T.J.C. Jacob (1984). ''Channel Recognition Using an Online Hardware Filter''. In Journal of Physiology, (London: Physiological Society), pp. 3–5</ref>), texture analysis(Southam et al, 2009<ref>Southam, P., and Harvey, R. (2009). ''Texture classification via morphological scale-space: Tex-Mex features''. J. Electron. Imaging 18, 043007–043007</ref>), lipreading(Matthews et al., 2002<ref>Matthews, I., Cootes, T.F., Bangham, J.A., Cox, S., and Harvey, R. (2002). ''Extraction of visual features for lipreading''. Pattern Anal. Mach. Intell. Ieee Trans. 24, 198–213</ref>). In 2D for segmenting 2D through extremal trees(Bangham et al., 1998<ref>Bangham, J.A., Hidalgo, J.R., Harvey, R., and Cawley, G. (1998). ''The segmentation of images via scale-space trees''. In Proceedings of British Machine Vision Conference, pp. 33–43</ref>), maximally stable contours(Lan et al., 2010<ref>Lan, Y., Harvey, R., and Perez Torres, J.R. (2010). ''Finding stable salient contours.'' Image Vis. Comput. 28, 1244–1254</ref>), images (), creating painterly pictures from photos(Bangham et al., 2003<ref>Bangham, J.A., Gibson, S.E., and Harvey, R. (2003). T''he art of scale-space''. In Proc. British Machine Vision Conference, pp. 569–578</ref>); and in 3D for segmenting volumes.
| | [http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software#Art.2C_extrema_of_light_and_shade:_PhotoArtMaster <span style="color:Navy;">'''MORE'''</span>] |
|
| |
|
| | ==[[Software#Reaction-diffusion and morphogenesis| <span style="color:Navy;"> '''Reaction-diffusion'''</span>]] <span style="color:Navy;">and morphogenesis - the growth of shapes== |
| | {| border="0" width=100% style="background-color:#000000;" |
| | |- |
| | |align="center"| |
| | [[Image:tentacles_reaction_diffusion.png|400px]] |
| | [[Image:tentacles_morphogenesis.png|600px]] |
| | |} |
| | This image forms part of a 'journey' in the Science Museum of London's 'Journeys of Invention' [http://www.sciencemuseum.org.uk/journeys iPad app.]<br><br> |
| | [[Software#Reaction-diffusion and morphogenesis|<span style="color:Navy;">'''MORE'''</span>]]<br><br> |
|
| |
|
| | | =[[Andrew personal | Andrew outside activities]]<br> |
| MSER's are used for finding objects in images, other sieves have been used in many ways, e.g. for lipreading<ref> ref name="Matthews"</ref>, for segmenting 2D and 3D images (maximally stable contours), texture analysis, analysing ID protein hydrophobicity plots, denoising single channel current data and creating painterly pictures from photos.
| |
| | |
| Art created using ArtMaster was featured in an exhibit at the London Victoria and Albert (V&A) Museum exhibition '???' exhibition [http://www.sciencemuseum.org.uk/visitmuseum/galleries/turing.aspx finding its name]. <br>
| |
| [[Software#MSERs extrema connected-set filters and sieves|<span style="color:Navy;">More details on MSERs, extrema, connected-set filters and sieves</span>]]<br><br> | |
| | |
| | |
| According to scientists, the Sun is pretty big <ref>E. Miller, The Sun, (New York: Academic Press, 2005), 23-5.</ref>.
| |
| In fact, it is very big <ref group="footnotes">Take their word for it. Don't look directly at the sun!</ref>.
| |
| | |
| ==Notes==
| |
| <references group="footnotes" />
| |
| ==References==
| |
| <references />
| |