Tutorial on the basic interaction function: Difference between revisions
No edit summary |
No edit summary |
||
| (14 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[GFtbox_Tutorial_pages|Back to tutorial pages]] | [[GFtbox_Tutorial_pages#2 Modelling using a combination of GUI and interaction function|Back to tutorial pages]] | ||
=Creating an interaction function= | |||
The goal is to re-create the model used to illustrate [[GFtbox#What does GFtbox require?|why we use matlab]]. We will create two new variables A and B - in the interaction function they are labelled'' id_a_p'' and ''id_b_p'' (why?[[Tutorial on the basic interaction function#Variables and utility functions| see below]]). The ''x,y,z'' coordinates of mesh nodes (vertices) are stored in the Mesh data structure in a field called ''nodes''. Thus the ''x'' coordinate of node 1 is stored in ''m.nodes(1,1)'', the ''y'' coordinate in ''m.nodes(1,2)'' and ''z'' coordinate in ''m.nodes(1,3)''. | |||
The patterns of morphogens A and B are set up by <br> | |||
id_a_p(m.nodes(:,1)<-0.03)=1; | |||
id_b_p(m.nodes(:,2)<-0.01)=1; | |||
where ''id_a_p'' is the A morphogen. ''m.node(:,1)'' refers to the ''x'' coordinates of all nodes (vertices) in the mesh<br> | |||
The expression ''(m.nodes(:,1)<-0.03)'' means find all vertices with ''x'' coordinates that are less than -0.03.<br><br> | |||
Similarly, ''(m.nodes(:,2)<-0.01)'' means find all vertices with ''y'' coordinates that are less than -0.01.<br><br> | |||
The model is articulated in the following code. | |||
if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code. | |||
id_a_p(m.nodes(:,1)<-0.03)=1; % setup region for A where identity factor A is represented by id_a_p | |||
id_b_p(m.nodes(:,2)<-0.01)=1; % setup region for B | |||
else | |||
% @@KRN Growth Regulatory Network | |||
kapar_p(:) = id_a_l .* inh(1,id_b_l); % growth rate | |||
kaper_p(:) = kapar_p; % isotropic growth | |||
kbpar_p(:) = kapar_p; % same on both sides of the sheet | |||
kbper_p(:) = kapar_p; % same | |||
knor_p(:) = 0; % thickness not growing | |||
end | |||
==Creating the interaction function from the GUI== | ==Creating the interaction function from the GUI== | ||
{| border="1" cellpadding="5" cellspacing="5" | {| border="1" cellpadding="5" cellspacing="5" | ||
|- valign="top" | |- valign="top" | ||
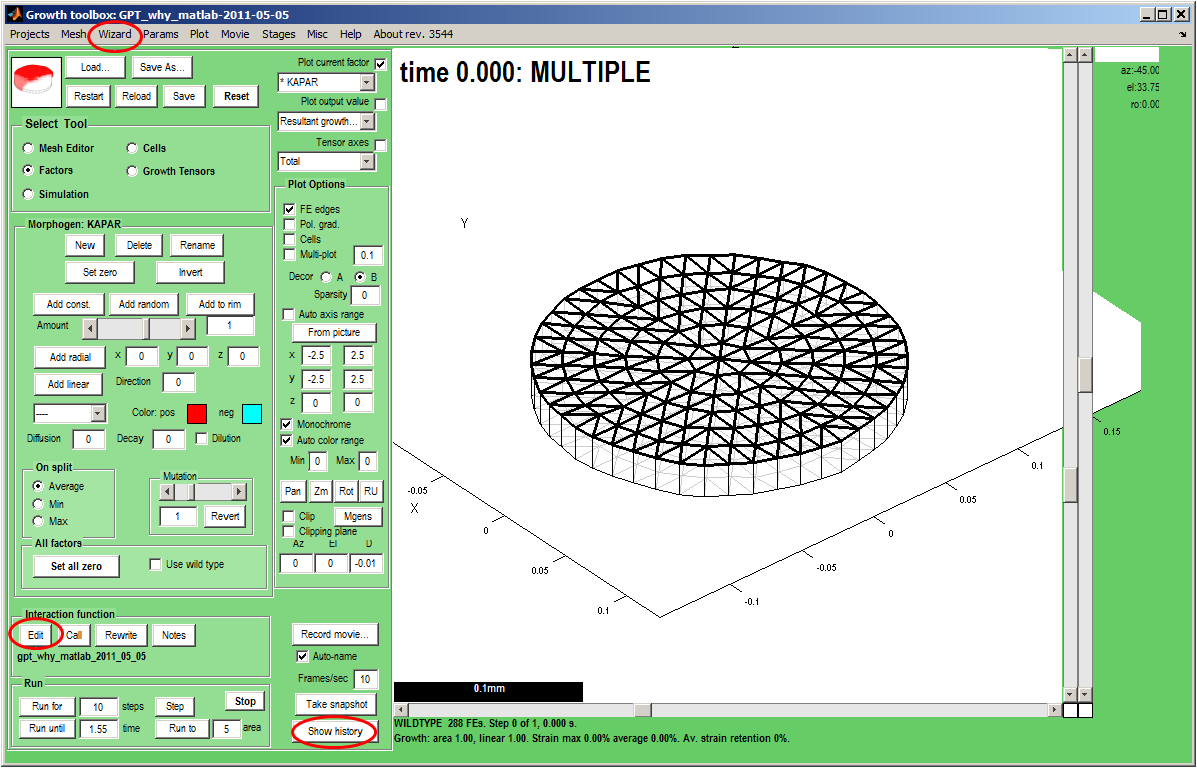
|width="300px"|The ''GFtbox'' user interface. | |width="300px"|The ''GFtbox'' user interface. Start a new project, with a disc shaped mesh and add two new morphogens ''id_a'' and ''id_b'' (Panel: Morphogens: New).<br><br> | ||
A free text file with the same name as the interaction function is also created automatically on pressing Panel:Interaction function: notes. This makes it easier to keep notes on what you are doing.<br><br> | Save As<br><br> | ||
Now create and edit the interaction function by clicking on (Panel: Interaction function: Edit) (bottom left) <br><br> | |||
There is lots of 'green stuff' - lines that are comments. Just for the moment we will more than ignore them - delete them. Look at the listing below to get an idea of what to delete. | |||
Now in Section 4 add the code shown above - it is shown in the listing below. | |||
<br><br> | |||
A free-text file with the same name as the interaction function is also created automatically on pressing Panel:Interaction function: notes. This makes it easier to keep notes on what you are doing.<br><br> | |||
<small>Note: you can see the internal data structure by clicking Menu: Wizard: Export mesh and follow the instructions shown at the command line.<br>Another way to see Mesh m is to set a break point in the interaction function and running (e.g. Panel: Run: Step, or Panel: Interaction function: Call, etc.)<br>Note: a history of commands that have been run recently can be displayed (can be used to help write custom modelling programs that exploit the GFtbox library) </small> | <small>Note: you can see the internal data structure by clicking Menu: Wizard: Export mesh and follow the instructions shown at the command line.<br>Another way to see Mesh m is to set a break point in the interaction function and running (e.g. Panel: Run: Step, or Panel: Interaction function: Call, etc.)<br>Note: a history of commands that have been run recently can be displayed (can be used to help write custom modelling programs that exploit the GFtbox library) </small> | ||
|width="500px"|[[File:Editwizardhistory.png|500px|GFtbox interface]] | |width="500px"|[[File:Editwizardhistory.png|500px|GFtbox interface]] | ||
| Line 42: | Line 71: | ||
|width="150pt"|Steps(m)|| current step number in Mesh m | |width="150pt"|Steps(m)|| current step number in Mesh m | ||
|- | |- | ||
|width="150pt"|pro(k,id)|| promote by k in regions designated by factor, id | |width="150pt"|pro(k,id)|| promote by k in regions designated by factor, id (''pro(k,'''x''')=1+k'''x'') | ||
|- | |- | ||
|width="150pt"|inh(k,id)|| inhibit by k in regions designated by factor, id | |width="150pt"|inh(k,id)|| inhibit by k in regions designated by factor, id (''inh(k,'''x''')=1/(1+k'''x''')'') | ||
|- | |- | ||
|width="150pt"|local_setproperties( m )|| initialise Mesh properties | |width="150pt"|local_setproperties( m )|| initialise Mesh properties | ||
| Line 52: | Line 81: | ||
|} | |} | ||
=The interaction function in more detail= | =The interaction function in more detail= | ||
A large number of comments have been deleted to allow us to focus on the essential components. (What are these deleted comments? They provide hints and clues on how to program a model - many people use the system and we found the easiest way for people to remember how to program models is to provide comments in the appropriate place - which can be deleted when not needed. Ok so you want see an uncensored version, [[Full freshly minted interaction function| here is a full freshly minted interaction function]]. [[Tutorial on the interaction function details|It is discussed in more detail here.]])<br><br> | |||
<span style="color: CornflowerBlue">'''This part is generated automatically from the GUI'''<br> | |||
The name matches the project name (the project name is forced into lowercase), GFtbox revision number refers to | |||
our source code control, lines starting with % are comments</span> | |||
function m = gpt_why_matlab_2011_05_05( m ) | |||
<span style="color: Green">%m = gpt_why_matlab_2011_05_05( m )</span> | |||
<span style="color: Green">% Morphogen interaction function.</span> | |||
<span style="color: Green">% Written at 2011-05-28 08:08:11.</span> | |||
<span style="color: Green">% GFtbox revision 3544, 2011-05-25 13:37:23.907904.</span> | |||
<span style="color: Green">% The user may edit any part of this function between delimiters</span> | |||
<span style="color: Green">% of the form "USER CODE..." and "END OF USER CODE...". The</span> | |||
<span style="color: Green">% delimiters themselves must not be moved, edited, deleted, or added.</span> | |||
if isempty(m), return; end | |||
fprintf( 1, '<span style="color: Green">%s found in %s\n', mfilename(), which(mfilename()) );</span> | |||
try | |||
m = local_setproperties( m ); | |||
catch | |||
end | |||
realtime = m.globalDynamicProps.currenttime; | |||
<span style="color: | <span style="color: Green">%%% USER CODE: INITIALISATION</span> | ||
<span style="color: CornflowerBlue">Section 2 This part could contain user code | |||
but in this case there is nothing we need to do before the system extracts information from the Mesh data structure</span> | |||
<span style="color: Green">%%% END OF USER CODE: INITIALISATION</span> | |||
<span style="color: CornflowerBlue">Section 3 This part is generated automatically from the GUI | |||
Variables are extracted from Mesh and the mesh itself is summarised in a set of comments</span> | |||
<span style="color: Green">%%% SECTION 1: ACCESSING MORPHOGENS AND TIME.</span> | |||
<span style="color: Green">%%% AUTOMATICALLY GENERATED CODE: DO NOT EDIT.</span> | |||
if isempty(m), return; end | |||
setGlobals(); | |||
global gNEW_KA_PAR gNEW_KA_PER gNEW_KB_PAR gNEW_KB_PER | |||
global gNEW_K_NOR gNEW_POLARISER gNEW_STRAINRET gNEW_ARREST | |||
dt = m.globalProps.timestep; | |||
polariser_i = gNEW_POLARISER; | |||
P = m.morphogens(:,polariser_i); | |||
[kapar_i,kapar_p,kapar_a,kapar_l] = getMgenLevels( m, 'KAPAR' ); | |||
[kaper_i,kaper_p,kaper_a,kaper_l] = getMgenLevels( m, 'KAPER' ); | |||
[kbpar_i,kbpar_p,kbpar_a,kbpar_l] = getMgenLevels( m, 'KBPAR' ); | |||
[kbper_i,kbper_p,kbper_a,kbper_l] = getMgenLevels( m, 'KBPER' ); | |||
[knor_i,knor_p,knor_a,knor_l] = getMgenLevels( m, 'KNOR' ); | |||
[strainret_i,strainret_p,strainret_a,strainret_l] = getMgenLevels( m, 'STRAINRET' ); | |||
[arrest_i,arrest_p,arrest_a,arrest_l] = getMgenLevels( m, 'ARREST' ); | |||
[id_a_i,id_a_p,id_a_a,id_a_l] = getMgenLevels( m, 'ID_A' ); | |||
[id_b_i,id_b_p,id_b_a,id_b_l] = getMgenLevels( m, 'ID_B' ); | |||
<span style="color: Green">% Mesh type: circle</span> | |||
<span style="color: Green">% centre: 0</span> | |||
<span style="color: Green">% circumpts: 48</span> | |||
<span style="color: Green">% coneangle: 0</span> | |||
<span style="color: Green">% dealign: 0</span> | |||
<span style="color: Green">% height: 0</span> | |||
<span style="color: Green">% innerpts: 0</span> | |||
<span style="color: Green">% randomness: 0.1</span> | |||
<span style="color: Green">% rings: 6</span> | |||
<span style="color: Green">% version: 1</span> | |||
<span style="color: Green">% xwidth: 0.2</span> | |||
<span style="color: Green">% ywidth: 0.2</span> | |||
<span style="color: Green">% Morphogen Diffusion Decay Dilution Mutant</span> | |||
<span style="color: Green">% -------------------------------------------------</span> | |||
<span style="color: Green">% KAPAR ---- ---- ---- ----</span> | |||
<span style="color: Green">% KAPER ---- ---- ---- ----</span> | |||
<span style="color: Green">% KBPAR ---- ---- ---- ----</span> | |||
<span style="color: Green">% KBPER ---- ---- ---- ----</span> | |||
</span> | <span style="color: Green">% KNOR ---- ---- ---- ----</span> | ||
<span style="color: Green">% POLARISER ---- ---- ---- ----</span> | |||
<span style="color: Green">% STRAINRET ---- ---- ---- ----</span> | |||
<span style="color: Green">% ARREST ---- ---- ---- ----</span> | |||
<span style="color: Green">% ID_A ---- ---- ---- ----</span> | |||
<span style="color: Green">% ID_B ---- ---- ---- ----</span> | |||
<span style="color: CornflowerBlue">Section 4 This part contains user code that articulates the model</span> | |||
<span style="color: Green">%%% USER CODE: MORPHOGEN INTERACTIONS</span> | |||
if (Steps(m)==0) && m.globalDynamicProps.doinit <span style="color: Green">% Initialisation code.</span> | |||
id_a_p(m.nodes(:,1)<-0.03)=1; <span style="color: Green">% setup region for A</span> | |||
id_b_p(m.nodes(:,2)<-0.01)=1; <span style="color: Green">% setup region for B</span> | |||
else | |||
<span style="color: Green">% @@KRN Growth Regulatory Network</span> | |||
kapar_p(:) = id_a_l .* inh(1,id_b_l); <span style="color: Green">% growth rate</span> | |||
kaper_p(:) = kapar_p; <span style="color: Green">% isotropic growth</span> | |||
kbpar_p(:) = kapar_p; <span style="color: Green">% same on both sides of the sheet</span> | |||
kbper_p(:) = kapar_p; <span style="color: Green">% same</span> | |||
knor_p(:) = 0; <span style="color: Green">% thickness not growing</span> | |||
end | |||
<span style="color: CornflowerBlue">Section 5 This part is generated automatically from the GUI | |||
Variables are put back into the data structure (Mesh). | |||
Note: only variables with the _p suffix (promotors) are stored (levels _l can be computed on the fly and activities _a are | |||
generally set in the GUI - they can only be changed in the interaction function by altering the data structure itself.</span> | |||
<span style="color: Green">%%% END OF USER CODE: MORPHOGEN INTERACTIONS</span> | |||
<span style="color: Green">%%% SECTION 6: INSTALLING MODIFIED VALUES BACK INTO MESH STRUCTURE</span> | |||
<span style="color: Green">%%% AUTOMATICALLY GENERATED CODE: DO NOT EDIT.</span> | |||
m.morphogens(:,polariser_i) = P; | |||
m.morphogens(:,kapar_i) = kapar_p; | |||
<span style="color: | m.morphogens(:,kaper_i) = kaper_p; | ||
m.morphogens(:,kbpar_i) = kbpar_p; | |||
m.morphogens(:,kbper_i) = kbper_p; | |||
<span style="color: CornflowerBlue"> | m.morphogens(:,knor_i) = knor_p; | ||
m.morphogens(:,strainret_i) = strainret_p; | |||
m.morphogens(:,arrest_i) = arrest_p; | |||
m.morphogens(:,id_a_i) = id_a_p; | |||
<span style="color: | m.morphogens(:,id_b_i) = id_b_p; | ||
<span style="color: Green">%%% USER CODE: FINALISATION</span> | |||
<span style="color: Green">%%% END OF USER CODE: FINALISATION</span> | |||
end | |||
<span style="color: Green">%%% USER CODE: SUBFUNCTIONS</span> | |||
<span style="color: CornflowerBlue">Section 5</span> | |||
function m = local_setproperties( m ) | |||
end | |||
Latest revision as of 17:36, 6 June 2011
Creating an interaction function
The goal is to re-create the model used to illustrate why we use matlab. We will create two new variables A and B - in the interaction function they are labelled id_a_p and id_b_p (why? see below). The x,y,z coordinates of mesh nodes (vertices) are stored in the Mesh data structure in a field called nodes. Thus the x coordinate of node 1 is stored in m.nodes(1,1), the y coordinate in m.nodes(1,2) and z coordinate in m.nodes(1,3).
The patterns of morphogens A and B are set up by
id_a_p(m.nodes(:,1)<-0.03)=1;
id_b_p(m.nodes(:,2)<-0.01)=1;
where id_a_p is the A morphogen. m.node(:,1) refers to the x coordinates of all nodes (vertices) in the mesh
The expression (m.nodes(:,1)<-0.03) means find all vertices with x coordinates that are less than -0.03.
Similarly, (m.nodes(:,2)<-0.01) means find all vertices with y coordinates that are less than -0.01.
The model is articulated in the following code.
if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code.
id_a_p(m.nodes(:,1)<-0.03)=1; % setup region for A where identity factor A is represented by id_a_p
id_b_p(m.nodes(:,2)<-0.01)=1; % setup region for B
else
% @@KRN Growth Regulatory Network
kapar_p(:) = id_a_l .* inh(1,id_b_l); % growth rate
kaper_p(:) = kapar_p; % isotropic growth
kbpar_p(:) = kapar_p; % same on both sides of the sheet
kbper_p(:) = kapar_p; % same
knor_p(:) = 0; % thickness not growing
end
Creating the interaction function from the GUI
Variables and utility functions
Variables that need to be seen in the GUI (morphogens, growth factors, signals, etc.) are always set up (declared) from the GUI using Morphogen Panel. We have evolved a naming convention
| prefixes |
| ||||||||
| suffixes | _p, _l, _a, _i |
Full details of types of morphogens and factors
| Pre-defined variables |
| ||||||||||
| Pre-defined functions |
|
The interaction function in more detail
A large number of comments have been deleted to allow us to focus on the essential components. (What are these deleted comments? They provide hints and clues on how to program a model - many people use the system and we found the easiest way for people to remember how to program models is to provide comments in the appropriate place - which can be deleted when not needed. Ok so you want see an uncensored version, here is a full freshly minted interaction function. It is discussed in more detail here.)
This part is generated automatically from the GUI
The name matches the project name (the project name is forced into lowercase), GFtbox revision number refers to our source code control, lines starting with % are comments function m = gpt_why_matlab_2011_05_05( m ) %m = gpt_why_matlab_2011_05_05( m ) % Morphogen interaction function. % Written at 2011-05-28 08:08:11. % GFtbox revision 3544, 2011-05-25 13:37:23.907904. % The user may edit any part of this function between delimiters % of the form "USER CODE..." and "END OF USER CODE...". The % delimiters themselves must not be moved, edited, deleted, or added. if isempty(m), return; end fprintf( 1, '%s found in %s\n', mfilename(), which(mfilename()) ); try m = local_setproperties( m ); catch end realtime = m.globalDynamicProps.currenttime; %%% USER CODE: INITIALISATION Section 2 This part could contain user code but in this case there is nothing we need to do before the system extracts information from the Mesh data structure %%% END OF USER CODE: INITIALISATION Section 3 This part is generated automatically from the GUI Variables are extracted from Mesh and the mesh itself is summarised in a set of comments %%% SECTION 1: ACCESSING MORPHOGENS AND TIME. %%% AUTOMATICALLY GENERATED CODE: DO NOT EDIT. if isempty(m), return; end setGlobals(); global gNEW_KA_PAR gNEW_KA_PER gNEW_KB_PAR gNEW_KB_PER global gNEW_K_NOR gNEW_POLARISER gNEW_STRAINRET gNEW_ARREST dt = m.globalProps.timestep; polariser_i = gNEW_POLARISER; P = m.morphogens(:,polariser_i); [kapar_i,kapar_p,kapar_a,kapar_l] = getMgenLevels( m, 'KAPAR' ); [kaper_i,kaper_p,kaper_a,kaper_l] = getMgenLevels( m, 'KAPER' ); [kbpar_i,kbpar_p,kbpar_a,kbpar_l] = getMgenLevels( m, 'KBPAR' ); [kbper_i,kbper_p,kbper_a,kbper_l] = getMgenLevels( m, 'KBPER' ); [knor_i,knor_p,knor_a,knor_l] = getMgenLevels( m, 'KNOR' ); [strainret_i,strainret_p,strainret_a,strainret_l] = getMgenLevels( m, 'STRAINRET' ); [arrest_i,arrest_p,arrest_a,arrest_l] = getMgenLevels( m, 'ARREST' ); [id_a_i,id_a_p,id_a_a,id_a_l] = getMgenLevels( m, 'ID_A' ); [id_b_i,id_b_p,id_b_a,id_b_l] = getMgenLevels( m, 'ID_B' ); % Mesh type: circle % centre: 0 % circumpts: 48 % coneangle: 0 % dealign: 0 % height: 0 % innerpts: 0 % randomness: 0.1 % rings: 6 % version: 1 % xwidth: 0.2 % ywidth: 0.2 % Morphogen Diffusion Decay Dilution Mutant % ------------------------------------------------- % KAPAR ---- ---- ---- ---- % KAPER ---- ---- ---- ---- % KBPAR ---- ---- ---- ---- % KBPER ---- ---- ---- ---- % KNOR ---- ---- ---- ---- % POLARISER ---- ---- ---- ---- % STRAINRET ---- ---- ---- ---- % ARREST ---- ---- ---- ---- % ID_A ---- ---- ---- ---- % ID_B ---- ---- ---- ---- Section 4 This part contains user code that articulates the model %%% USER CODE: MORPHOGEN INTERACTIONS if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code. id_a_p(m.nodes(:,1)<-0.03)=1; % setup region for A id_b_p(m.nodes(:,2)<-0.01)=1; % setup region for B else % @@KRN Growth Regulatory Network kapar_p(:) = id_a_l .* inh(1,id_b_l); % growth rate kaper_p(:) = kapar_p; % isotropic growth kbpar_p(:) = kapar_p; % same on both sides of the sheet kbper_p(:) = kapar_p; % same knor_p(:) = 0; % thickness not growing end Section 5 This part is generated automatically from the GUI Variables are put back into the data structure (Mesh). Note: only variables with the _p suffix (promotors) are stored (levels _l can be computed on the fly and activities _a are generally set in the GUI - they can only be changed in the interaction function by altering the data structure itself. %%% END OF USER CODE: MORPHOGEN INTERACTIONS %%% SECTION 6: INSTALLING MODIFIED VALUES BACK INTO MESH STRUCTURE %%% AUTOMATICALLY GENERATED CODE: DO NOT EDIT. m.morphogens(:,polariser_i) = P; m.morphogens(:,kapar_i) = kapar_p; m.morphogens(:,kaper_i) = kaper_p; m.morphogens(:,kbpar_i) = kbpar_p; m.morphogens(:,kbper_i) = kbper_p; m.morphogens(:,knor_i) = knor_p; m.morphogens(:,strainret_i) = strainret_p; m.morphogens(:,arrest_i) = arrest_p; m.morphogens(:,id_a_i) = id_a_p; m.morphogens(:,id_b_i) = id_b_p; %%% USER CODE: FINALISATION %%% END OF USER CODE: FINALISATION end %%% USER CODE: SUBFUNCTIONS Section 5 function m = local_setproperties( m ) end