"Visualising Plant Development and Gene Expression in 3D using Optical Projection Tomography": Difference between revisions
No edit summary |
No edit summary |
||
| (53 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Image:Plant cell.png|right|thumb|256px|Top row; volume view of mature ATHB8:GUS Arabidopsis leaf with stained veins (m2, 9 dfs) displayed using QtVolView lighting and tone-shader effects. Middle row; combined transmission and fluorescent (GFP1) OPT channels. Visible channel is red, fluorescent channel is green. Bottom row; stained veins extracted using semi-automatic segmentation tools. In the application the leaf can be viewed from any angle, re-coloured, etc.]] | |||
=Abstract= | |||
A deeper understanding of the mechanisms that underlie plant growth and development requires quantitative data on three-dimensional (3D) morphology and gene activity at a variety of stages and scales. To address this, we have explored the use of optical projection tomography (OPT) as a method for capturing 3D data from plant specimens. We show that OPT can be conveniently applied to a wide variety of plant material at a range of scales, including seedlings, leaves, flowers, roots, seeds, embryos, and meristems. At the highest resolution, large individual cells can be seen in the context of the surrounding plant structure. For naturally semitransparent structures, such as roots, live 3D imaging using OPT is also possible. 3D domains of gene expression can be visualized using either marker genes, such as b-glucuronidase, or more directly by whole-mount in situ hybridization. We also describe tools and software that allow the 3D data to be readily quantified and visualized interactively in different ways. | |||
=Manuscript= | =Manuscript= | ||
[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/VisualisingPlantDevelopmentandGeneExpressionin3DusingOpticalProjectionTomography.pdf "Visualising Plant Development and Gene Expression in 3D using Optical Projection Tomography"], ''K. Lee, J. Avondo, H. Morrison, L. Blot, M. Stark, J. Sharpe, J. A. Bangham, and E. S. Coen'' | [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/VisualisingPlantDevelopmentandGeneExpressionin3DusingOpticalProjectionTomography.pdf "Visualising Plant Development and Gene Expression in 3D using Optical Projection Tomography"], ''K. Lee, J. Avondo, H. Morrison, L. Blot, M. Stark, J. Sharpe, J. A. Bangham, and E. S. Coen'' | ||
=Sumplementary Material= | |||
==Software== | |||
The software used to produce all the figures in the paper can be found below. | |||
===QtVolViewerLITE v1=== | |||
The QtVolViewLITE program is designed to run on any PC with a 64 Mbyte graphic card. | |||
[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/QtVolViewerLITEv1.zip QtVolViewerLITEv1.zip] | [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/QtVolViewerLITEv1QuickStart.pdf QuickStart.pdf] | [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/QtVolViewerLITEv1UserManual.pdf UserManual.pdf] | |||
===QtVolViewer v1.64=== | |||
Latest public version of QtVolViewer. Only tested on the following hardware: Nvidia 7900GTX & Nvidia 8800GTX. This version uses advanced features of OpenGL such as the OpenGL Shanding Language (GLSL) and Frame Buffer Objects. | |||
[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/QtVolviewerV1.64.zip QtVolviewerV1.64.zip] | |||
===MovieMake=== | |||
This software is useful to make movies out of stack of images. See the PDF included in the zip for instructions. | |||
[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/MovieMakev1_1.zip MovieMakev1_1.zip] | |||
===WLZ to PNG Converter=== | |||
This software is useful to convert WLZ files to stacks of PNGs. Put your WLZ in the IN directory, then run the program and the stacks will appear in the OUT directory as subdirectories for each WLZ filename. | |||
[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/QtVolProcess_rev1396.zip QtVolProcess_rev1396.zip] | |||
==Datasets used in Figures== | |||
Below you can find the actual PNG stacks for the datasets presented in the paper. To download a specific dataset click on the DOWNLOAD link below the animated GIF preview of the dataset. | |||
===Single channel data sets:=== | |||
{| border="0" cellspacing="10" cellpadding="5" | |||
|- | |||
| align="center"| [[Image:Figure1.gif]] || [[Image:Figure2.gif]] || [[Image:Figure3.gif]] || [[Image:Figure4.gif]] | |||
|- | |||
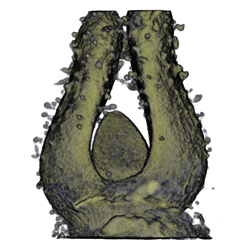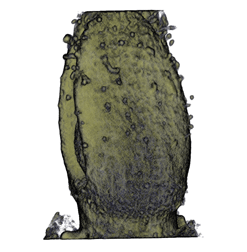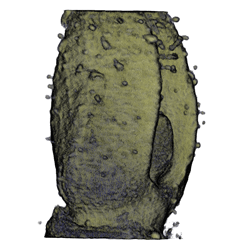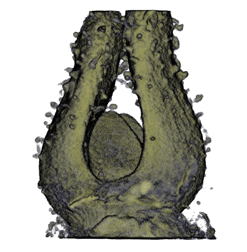
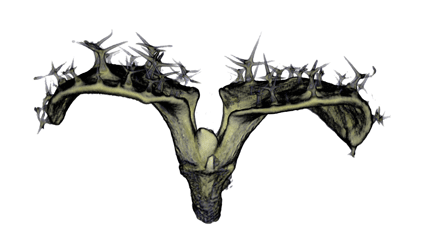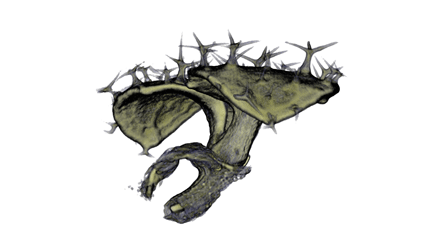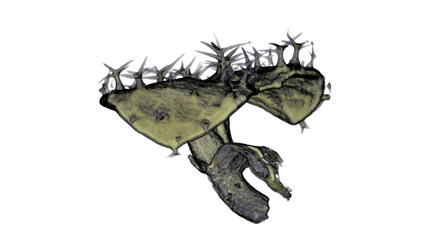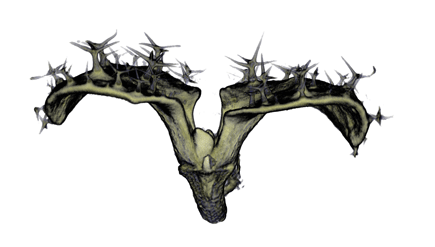
| align="center"| '''Antirinhium Flower''' || align="center"| '''Antirinhium Meristem''' || align="center"| '''Arabidopsis Seedling''' || align="center"| '''Arabidopsis Slique''' | |||
|- | |||
| align="center"| [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/PlantCellOPT/Anti_Flower294.zip Download] || align="center"| [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/PlantCellOPT/Antiriniuhm_Meristem(r512g110usmall).zip Download] || align="center"| [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/PlantCellOPT/Arab_Seedling(174).zip Download]|| align="center"|[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/PlantCellOPT/Arabidopsis_Slique(372).zip Download] | |||
|} | |||
===Multi channel data sets:=== | |||
{| border="0" cellspacing="10" cellpadding="5" | |||
|- | |||
| align="center"| [[Image:Figure5.gif]] || [[Image:Figure7.gif]] || [[Image:Figure8.gif]] || [[Image:Figure6.gif]] | |||
|- | |||
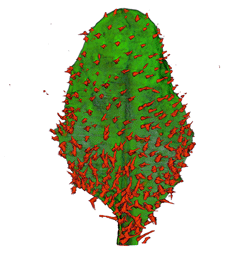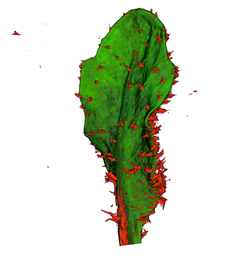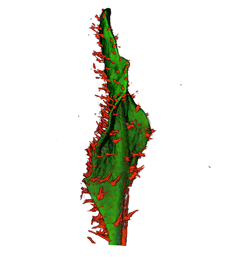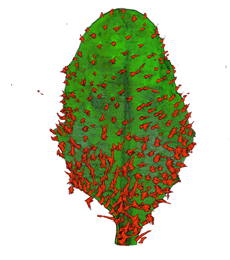
| align="center"| '''Arabidopsis Leaf''' (GL2:GUS expression in red) || align="center"| '''Arabidopsis Leaf''' (Ath8::GUS expression in red) || align="center"| '''Arabidopsis Meristem''' (Ath8:::GUS expression in red) || align="center"| '''Arabidopsis Meristem''' (LFY::GUS expression in red) | |||
|- | |||
| align="center"| [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/PlantCellOPT/Arab_LeafGL2_GUS(624).zip Download] || align="center"| [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/PlantCellOPT/ArabidopsisLeafAth8_GUS(460).zip Download] || align="center" | [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/PlantCellOPT/ArabidopsisSeedlingAth8_GUS(463).zip Download] || align="center"| [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/PlantCellOPT/Arabidopsis_MeristemLFY_GUS(423).zip Download] | |||
|} | |||
=Acknowledgements= | |||
BBSRC for grant support, JIC/UEA/MRC and MRC Technology. | |||
=Software team= | |||
Jerome Avondo with help from Lilian Blot and the Bangham group in the Computational Biology Group, Computing Sciences, University of East Anglia, Norwich, | |||
Latest revision as of 12:40, 12 June 2009

Abstract
A deeper understanding of the mechanisms that underlie plant growth and development requires quantitative data on three-dimensional (3D) morphology and gene activity at a variety of stages and scales. To address this, we have explored the use of optical projection tomography (OPT) as a method for capturing 3D data from plant specimens. We show that OPT can be conveniently applied to a wide variety of plant material at a range of scales, including seedlings, leaves, flowers, roots, seeds, embryos, and meristems. At the highest resolution, large individual cells can be seen in the context of the surrounding plant structure. For naturally semitransparent structures, such as roots, live 3D imaging using OPT is also possible. 3D domains of gene expression can be visualized using either marker genes, such as b-glucuronidase, or more directly by whole-mount in situ hybridization. We also describe tools and software that allow the 3D data to be readily quantified and visualized interactively in different ways.
Manuscript
"Visualising Plant Development and Gene Expression in 3D using Optical Projection Tomography", K. Lee, J. Avondo, H. Morrison, L. Blot, M. Stark, J. Sharpe, J. A. Bangham, and E. S. Coen
Sumplementary Material
Software
The software used to produce all the figures in the paper can be found below.
QtVolViewerLITE v1
The QtVolViewLITE program is designed to run on any PC with a 64 Mbyte graphic card.
QtVolViewerLITEv1.zip | QuickStart.pdf | UserManual.pdf
QtVolViewer v1.64
Latest public version of QtVolViewer. Only tested on the following hardware: Nvidia 7900GTX & Nvidia 8800GTX. This version uses advanced features of OpenGL such as the OpenGL Shanding Language (GLSL) and Frame Buffer Objects.
MovieMake
This software is useful to make movies out of stack of images. See the PDF included in the zip for instructions.
WLZ to PNG Converter
This software is useful to convert WLZ files to stacks of PNGs. Put your WLZ in the IN directory, then run the program and the stacks will appear in the OUT directory as subdirectories for each WLZ filename.
Datasets used in Figures
Below you can find the actual PNG stacks for the datasets presented in the paper. To download a specific dataset click on the DOWNLOAD link below the animated GIF preview of the dataset.
Single channel data sets:
 |
 |
 |

|
| Antirinhium Flower | Antirinhium Meristem | Arabidopsis Seedling | Arabidopsis Slique |
| Download | Download | Download | Download |
Multi channel data sets:
 |
 |
 |
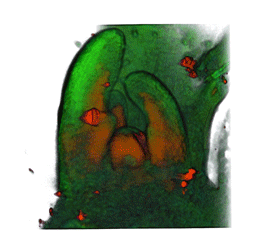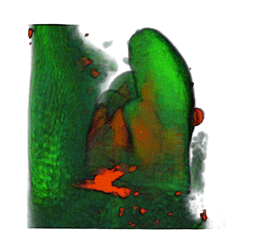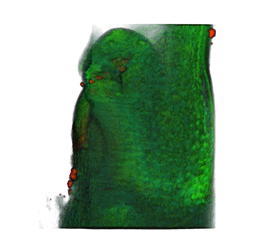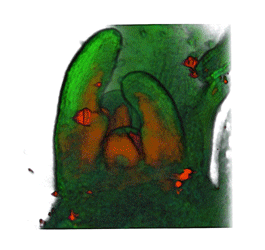
|
| Arabidopsis Leaf (GL2:GUS expression in red) | Arabidopsis Leaf (Ath8::GUS expression in red) | Arabidopsis Meristem (Ath8:::GUS expression in red) | Arabidopsis Meristem (LFY::GUS expression in red) |
| Download | Download | Download | Download |
Acknowledgements
BBSRC for grant support, JIC/UEA/MRC and MRC Technology.
Software team
Jerome Avondo with help from Lilian Blot and the Bangham group in the Computational Biology Group, Computing Sciences, University of East Anglia, Norwich,