"Visualising Plant Development and Gene Expression in 3D using Optical Projection Tomography"

Abstract
A deeper understanding of the mechanisms that underlie plant growth and development requires quantitative data on three-dimensional (3D) morphology and gene activity at a variety of stages and scales. To address this, we have explored the use of optical projection tomography (OPT) as a method for capturing 3D data from plant specimens. We show that OPT can be conveniently applied to a wide variety of plant material at a range of scales, including seedlings, leaves, flowers, roots, seeds, embryos, and meristems. At the highest resolution, large individual cells can be seen in the context of the surrounding plant structure. For naturally semitransparent structures, such as roots, live 3D imaging using OPT is also possible. 3D domains of gene expression can be visualized using either marker genes, such as b-glucuronidase, or more directly by whole-mount in situ hybridization. We also describe tools and software that allow the 3D data to be readily quantified and visualized interactively in different ways.
Manuscript
"Visualising Plant Development and Gene Expression in 3D using Optical Projection Tomography", K. Lee, J. Avondo, H. Morrison, L. Blot, M. Stark, J. Sharpe, J. A. Bangham, and E. S. Coen
Sumplementary Material
Software
QtVolViewerLITE v1
The QtVolViewLITE program is designed to run on any PC with a 64 Mbyte graphic card. It is suitable for viewing the data shown in Figures 1A and 2A as colour OPT volume and section views of the Antirrhinum flower. Internal floral structures such as anther lobes and the ovary at the base of the carpel are revealed. Three OPT scan channels are visible. Transmission (shown in blue), endogenous fluorescence, Leica TXR filter (shown in red) and GFP fluorescence, GFP1 filter (shown in green).
QtVolViewerLITEv1.zip | QuickStart.pdf | UserManual.pdf
QtVolViewer v1.64
Latest public version of QtVolViewer. Only tested on the following hardware: Nvidia 7900GTX & Nvidia 8800GTX. This version uses advanced features of OpenGL such as the OpenGL Shanding Language (GLSL) and Frame Buffer Objects.
Datasets used in Figures
Single channel data sets:
 |
 |
 |

|
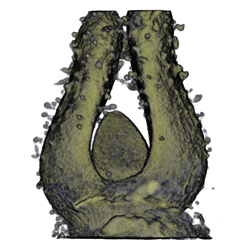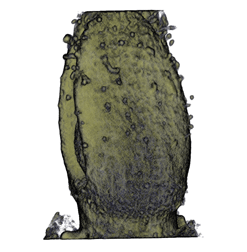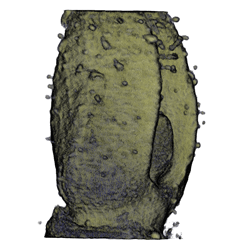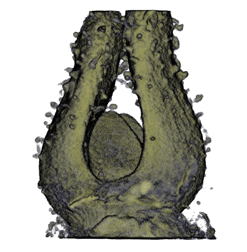
| Antirinhium Flower | Antirinhium Meristem | Arabidopsis Seedling | Arabidopsis Slique |
Multi channel data sets:
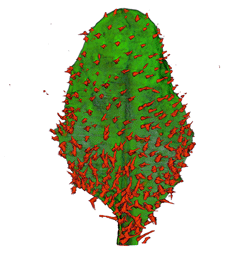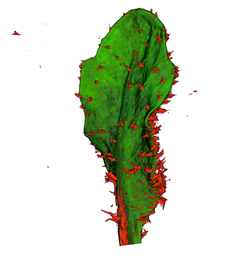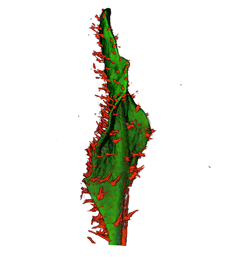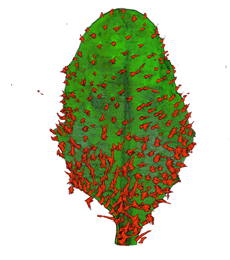 |
 |
 |
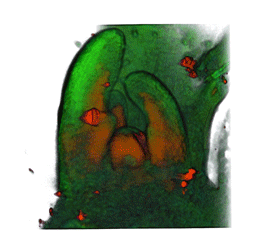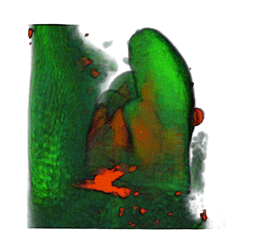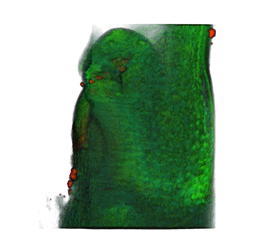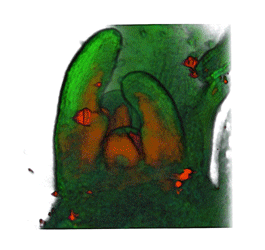
|
| Arabidopsis Leaf (GL2:GUS expression in red) | Arabidopsis Leaf (Ath8::GUS expression in red) | Arabidopsis Meristem (Ath8:::GUS expression in red) | Arabidopsis Meristem (LFY::GUS expression in red) |
Acknowledgements
BBSRC for grant support, JIC/UEA/MRC and MRC Technology.
Software team
Jerome Avondo with help from Lilian Blot and the Bangham group in the Computational Biology Group, Computing Sciences, University of East Anglia, Norwich,