"Mutational spaces for leaf shape and size": Difference between revisions
No edit summary |
No edit summary |
||
| (31 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== | =Abstract= | ||
A key approach to understanding how genes control growth and form is to analyze mutants in which shape and size have been perturbed. Although many | |||
mutants of this kind have been described in plants and animals, a general quantitative framework for describing them has yet to be established. Here we describe an approach based on Principal Component Analysis of organ landmarks and outlines. Applying this method to a collection of leaf shape mutants in Arabidopsis and Antirrhinum allows low-dimensional spaces to be constructed that capture the key variations in shape and size. Mutant phenotypes can be represented as vectors in these allometric spaces, allowing additive gene interactions to be readily described. The principal axis of each allometric space reflects size variation and an associated shape change. The shape change is similar to that observed during the later stages of normal development, suggesting that many phenotypic differences involve modulations in the timing of growth arrest. Comparison between allometric mutant spaces from different species reveals a similar range of phenotypic possibilities. The spaces therefore provide a general quantitative framework for exploring and comparing the development and evolution of form. | |||
[http:// | =The Manuscript= | ||
[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/MutationalSpacesForLeafShapeAndSize_Bensmihen.pdf “Mutational spaces for leaf shape and size”], ''S. Bensmihen, A. I. Hanna, N. B. Langlade, J. L. Micol, A. Bangham, and E. S. Coen'' | |||
=Downloading the Project and AAMToolbox= | |||
[[Image:HFSP.jpg|thumb||right]] | |||
The project used to create the figures in the manuscript, together with the Matlab toolbox that manages the project can be downloaded | |||
[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/papers/MutationalSpacesForLeafShapeAndSize.zip here]. ''(This contains version 6.5 of the AAMToolbox)'' | |||
To create the figures shown in the manuscript “Mutational spaces for leaf shape and size”, ''S. Bensmihen, A. I. Hanna, N. B. Langlade, J. L. Micol, A. Bangham, and E. S. Coen'', HFSP Journal, 2008, please follow the instructions below. | To create the figures shown in the manuscript “Mutational spaces for leaf shape and size”, ''S. Bensmihen, A. I. Hanna, N. B. Langlade, J. L. Micol, A. Bangham, and E. S. Coen'', HFSP Journal, 2008, please follow the instructions below. | ||
| Line 7: | Line 17: | ||
These figures were created using ''Matlab 2006b version 7.3.0.267'' running on ''Windows XP''. | These figures were created using ''Matlab 2006b version 7.3.0.267'' running on ''Windows XP''. | ||
=Installing the AAMToolbox= | |||
To install the toolbox, simply unzip the zip file you downloaded from the | |||
To install the toolbox, simply unzip the zip file you downloaded from the section above. In that folder you will find 2 folders, the first one will be the ‘AAMToolbox’ folder (this contains the Matlab files that run the toolbox). The second folder is named ‘PRJ_Arabid_Antirrh’. This is the project folder that contains all the data for the manuscript. To install the toolbox, from the Matlab prompt type | |||
<pre> | <pre> | ||
>>pathtool | >>pathtool | ||
</pre> | </pre> | ||
Click the ‘Add with subfolders’ button and then select the AAMToolbox folder. Save your new path and return to Matlab. You have now installed the toolbox. REMEMBER, ANY CHANGES YOU MAKE TO THE TOOLBOX DIRECTORY WILL REQUIRE YOU TO RESET THE PATH USING PATHOOL. | Click the ‘Add with subfolders’ button and then select the AAMToolbox folder. Save your new path and return to Matlab. You have now installed the toolbox. '''REMEMBER, ANY CHANGES YOU MAKE TO THE TOOLBOX DIRECTORY WILL REQUIRE YOU TO RESET THE PATH USING PATHOOL'''. | ||
=Creating Figures= | |||
==Figure 1== | |||
From the Matlab prompt type | From the Matlab prompt type | ||
<pre> | <pre> | ||
| Line 24: | Line 36: | ||
And select ''LE50template_04_05_06.temp_dat'' from the ''Templates'' directory. The colours are your choice. | And select ''LE50template_04_05_06.temp_dat'' from the ''Templates'' directory. The colours are your choice. | ||
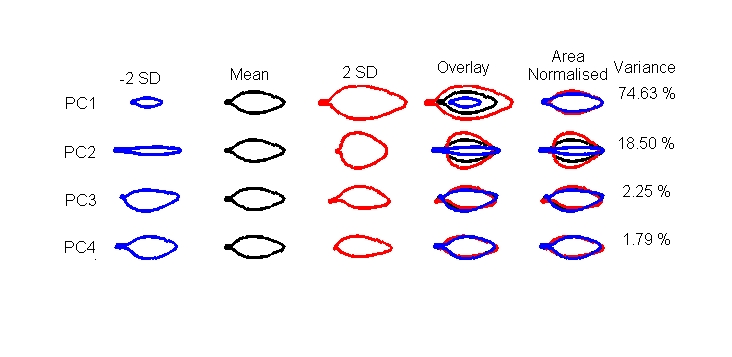
==Figure 2== | |||
From the Matlab prompt type | From the Matlab prompt type | ||
<pre> | <pre> | ||
| Line 34: | Line 47: | ||
* Next choose to use the mean from the model and finally select 2SD range. (The vertical and horizontal ranges can be changed but the default gets you started). | * Next choose to use the mean from the model and finally select 2SD range. (The vertical and horizontal ranges can be changed but the default gets you started). | ||
==Figure 3== | |||
From the Matlab prompt type | From the Matlab prompt type | ||
<pre> | <pre> | ||
| Line 47: | Line 61: | ||
* Next click the “Choose Shape Axes” button and select the PC 1. (To view another PC click the “Choose Shape Axes” button and choose the PC of interest (in our case PC1, PC2, PC3, PC4).) | * Next click the “Choose Shape Axes” button and select the PC 1. (To view another PC click the “Choose Shape Axes” button and choose the PC of interest (in our case PC1, PC2, PC3, PC4).) | ||
==Figure 4== | |||
'''UNDER CONSTRUCTION''' | '''UNDER CONSTRUCTION''' | ||
==Figure 5== | |||
From the Matlab prompt type | From the Matlab prompt type | ||
<pre> | <pre> | ||
| Line 59: | Line 75: | ||
* Next choose to use the mean from the model and finally select 2SD range. (The vertical and horizontal ranges can be changed but the default gets you started). | * Next choose to use the mean from the model and finally select 2SD range. (The vertical and horizontal ranges can be changed but the default gets you started). | ||
==Figure 6== | |||
From the Matlab prompt type | From the Matlab prompt type | ||
<pre> | <pre> | ||
| Line 68: | Line 85: | ||
* Next choose to use the mean from the model and finally select 2SD range. (The vertical and horizontal ranges can be changed but the default gets you started). | * Next choose to use the mean from the model and finally select 2SD range. (The vertical and horizontal ranges can be changed but the default gets you started). | ||
==Figure 7== | |||
'''UNDER CONSTRUCTION''' | '''UNDER CONSTRUCTION''' | ||
==Figure 8== | |||
From the Matlab prompt type | From the Matlab prompt type | ||
<pre> | <pre> | ||
| Line 85: | Line 104: | ||
(The leaf pictures originated from the original pictures (observed). Vectors were added in the Adobe Illustrator CS2 software) | (The leaf pictures originated from the original pictures (observed). Vectors were added in the Adobe Illustrator CS2 software) | ||
==Figure 9== | |||
* Top panel: the outlines of young and mature Arabidopsis and antirrhinum leaves from Yvette’s pictures (for Munderman et al.?) were cut and filled (using the magic stick from Adobe Photoshop). Pictures were copied to Adobe Illustrator CS2 and scaled (selecting your shape, using then from the bar menu: object, transformation, scaling (“mise à l’échelle” in the French version). | * Top panel: the outlines of young and mature Arabidopsis and antirrhinum leaves from Yvette’s pictures (for Munderman et al.?) were cut and filled (using the magic stick from Adobe Photoshop). Pictures were copied to Adobe Illustrator CS2 and scaled (selecting your shape, using then from the bar menu: object, transformation, scaling (“mise à l’échelle” in the French version). | ||
* Bottom panel: the outlines of the Arabidopsis and Antirrhinum mean shapes, plus or minus 2SD, were created using the figure_2_pc_effects script as described for Figure 2. The file was saved as .eps and opened in Adobe Illustrator CS2 and the same scaling procedure was used. | * Bottom panel: the outlines of the Arabidopsis and Antirrhinum mean shapes, plus or minus 2SD, were created using the figure_2_pc_effects script as described for Figure 2. The file was saved as .eps and opened in Adobe Illustrator CS2 and the same scaling procedure was used. | ||
=Creating Tables= | |||
* Table 1 originates from the ranked t-test analysis described above performed on all the Arabidopsis Ler groups (on Image List 18 model). Groups were arranged and sorted in XL. | * Table 1 originates from the ranked t-test analysis described above performed on all the Arabidopsis Ler groups (on Image List 18 model). Groups were arranged and sorted in XL. | ||
* Table 2 originates from the ranked t-test analysis (from Image List 21 model) of the antirrhinum groups (E_groups) using as wild type a group of 3 flower mutant groups (“flower_only_formean” group). | * Table 2 originates from the ranked t-test analysis (from Image List 21 model) of the antirrhinum groups (E_groups) using as wild type a group of 3 flower mutant groups (“flower_only_formean” group). | ||
Latest revision as of 11:48, 24 July 2008
Abstract
A key approach to understanding how genes control growth and form is to analyze mutants in which shape and size have been perturbed. Although many mutants of this kind have been described in plants and animals, a general quantitative framework for describing them has yet to be established. Here we describe an approach based on Principal Component Analysis of organ landmarks and outlines. Applying this method to a collection of leaf shape mutants in Arabidopsis and Antirrhinum allows low-dimensional spaces to be constructed that capture the key variations in shape and size. Mutant phenotypes can be represented as vectors in these allometric spaces, allowing additive gene interactions to be readily described. The principal axis of each allometric space reflects size variation and an associated shape change. The shape change is similar to that observed during the later stages of normal development, suggesting that many phenotypic differences involve modulations in the timing of growth arrest. Comparison between allometric mutant spaces from different species reveals a similar range of phenotypic possibilities. The spaces therefore provide a general quantitative framework for exploring and comparing the development and evolution of form.
The Manuscript
“Mutational spaces for leaf shape and size”, S. Bensmihen, A. I. Hanna, N. B. Langlade, J. L. Micol, A. Bangham, and E. S. Coen
Downloading the Project and AAMToolbox
The project used to create the figures in the manuscript, together with the Matlab toolbox that manages the project can be downloaded here. (This contains version 6.5 of the AAMToolbox)
To create the figures shown in the manuscript “Mutational spaces for leaf shape and size”, S. Bensmihen, A. I. Hanna, N. B. Langlade, J. L. Micol, A. Bangham, and E. S. Coen, HFSP Journal, 2008, please follow the instructions below.
These figures were created using Matlab 2006b version 7.3.0.267 running on Windows XP.
Installing the AAMToolbox
To install the toolbox, simply unzip the zip file you downloaded from the section above. In that folder you will find 2 folders, the first one will be the ‘AAMToolbox’ folder (this contains the Matlab files that run the toolbox). The second folder is named ‘PRJ_Arabid_Antirrh’. This is the project folder that contains all the data for the manuscript. To install the toolbox, from the Matlab prompt type
>>pathtool
Click the ‘Add with subfolders’ button and then select the AAMToolbox folder. Save your new path and return to Matlab. You have now installed the toolbox. REMEMBER, ANY CHANGES YOU MAKE TO THE TOOLBOX DIRECTORY WILL REQUIRE YOU TO RESET THE PATH USING PATHOOL.
Creating Figures
Figure 1
From the Matlab prompt type
>>figure_1_points_around_leaf
And select LE50template_04_05_06.temp_dat from the Templates directory. The colours are your choice.
Figure 2
From the Matlab prompt type
>>figure_2_pc_effects
- Select the folder ‘PRJ_Arabid_Antirrh\StatisticalModels\LE50Template_04_05_06\Set_1\ImageList_18’ for model (Ler only, not scaled).
- Next choose the PCs you want to display (1 to 5 in our case).
- Next choose to use the mean from the model and finally select 2SD range. (The vertical and horizontal ranges can be changed but the default gets you started).
Figure 3
From the Matlab prompt type
>>AAMToolbox
- Make sure that the current model is “LE50Template_04_05_06\Set_1\ImageList_18” from the drop down list.
- Next click the “View Shape Space” button.
- Next click the “Open” button from the Groups panel and select “all_groups_no_tsk”.
- Next click the “Load” button from the Groups panel.
- Next select ‘Tools-Units-Standard Deviations’.
- Next uncheck the “Group Labels” from the plot options panel.
- Next click the “Choose Shape Axes” button and select the PC 1. (To view another PC click the “Choose Shape Axes” button and choose the PC of interest (in our case PC1, PC2, PC3, PC4).)
Figure 4
UNDER CONSTRUCTION
Figure 5
From the Matlab prompt type
>>figure_2_pc_effects
- Select the folder ‘PRJ_Arabid_Antirrh\StatisticalModels\LE50Template_04_05_06\Set_1\ImageList_17’ for model (Ler only, scaled).
- Next choose the PCs you want to display (1 to 4 in our case).
- Next choose to use the mean from the model and finally select 2SD range. (The vertical and horizontal ranges can be changed but the default gets you started).
Figure 6
From the Matlab prompt type
>>figure_2_pc_effects
- Select the folder ‘PRJ_Arabid_Antirrh\StatisticalModels\LE50Template_04_05_06\Set_1\ImageList_21’ for model (_epure= cleaned form double mutant and flower effect mutants).
- Next choose the PCs you want to display (1 to 4 in our case).
- Next choose to use the mean from the model and finally select 2SD range. (The vertical and horizontal ranges can be changed but the default gets you started).
Figure 7
UNDER CONSTRUCTION
Figure 8
From the Matlab prompt type
>>AAMToolbox
- Make sure that the current model is “LE50Template LE50Template_04_05_06\Set_1\ImageList_21” from the drop down list.
- Next click the “View Shape Space” button.
- Next click the “Open” button from the Groups panel and select “ran_re _double”.
- Next click the “Load” button from the Groups panel.
- Next click the “Choose Shape Axes” button.
- Next select PCs 1 and 2 and hit “Ok”
- Next uncheck “Particles” and “Group Labels” from the “Plot Options” panel.
(The leaf pictures originated from the original pictures (observed). Vectors were added in the Adobe Illustrator CS2 software)
Figure 9
- Top panel: the outlines of young and mature Arabidopsis and antirrhinum leaves from Yvette’s pictures (for Munderman et al.?) were cut and filled (using the magic stick from Adobe Photoshop). Pictures were copied to Adobe Illustrator CS2 and scaled (selecting your shape, using then from the bar menu: object, transformation, scaling (“mise à l’échelle” in the French version).
- Bottom panel: the outlines of the Arabidopsis and Antirrhinum mean shapes, plus or minus 2SD, were created using the figure_2_pc_effects script as described for Figure 2. The file was saved as .eps and opened in Adobe Illustrator CS2 and the same scaling procedure was used.
Creating Tables
- Table 1 originates from the ranked t-test analysis described above performed on all the Arabidopsis Ler groups (on Image List 18 model). Groups were arranged and sorted in XL.
- Table 2 originates from the ranked t-test analysis (from Image List 21 model) of the antirrhinum groups (E_groups) using as wild type a group of 3 flower mutant groups (“flower_only_formean” group).